Abstract
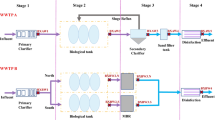
Aquaponics is defined as a sustainable and integrated system that combines fish aquaculture and hydroponic plant production in the same recirculated water loop. A recent study using high-throughput sequencing (HTS) technologies highlighted that microbial communities from an aquaponic system could control one of the most problematic pathogens in soilless lettuce culture, namely, Pythium aphanidermatum. Therefore, this study aims at isolating the microorganisms responsible for this biocontrol action. Based on the most promising genera identified by HTS, an innovative strategy for isolating and testing original biocontrol agents from aquaponic water was designed to control P. aphanidermatum. Eighty-two bacterial strains and 18 fungal strains were isolated, identified by Sanger sequencing, and screened in vivo to control damping-off of lettuce seeds caused by P. aphanidermatum. Out of these 100 isolates, the eight most efficacious ones were selected and further tested individually to control root rot disease caused by the same pathogen at a later stage of lettuce growth. Strains SHb30 (Sphingobium xenophagum), G2 (Aspergillus flavus), and Chito13 (Mycolicibacterium fortuitum) decreased seed damping-off at a better rate than a propamocarb fungicide and a Pseudomonas chlororaphis registered biocontrol agent did. In root rot bioassays, lettuce mortality was prevented by applying strains G2 and Chito13, which were at least as efficacious as the fungicide or biopesticide controls. Lettuce disease symptoms and mortality were eradicated by strain SHb30 in the first bioassay, but not in the second one. These results show that aquaponic systems are promising sources of original biocontrol agents, and that HTS-guided strategies could represent interesting approaches to identify new biocontrol agents.


Similar content being viewed by others
Data Availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Vallance J, Déniel F, Le Floch G et al (2010) Pathogenic and beneficial microorganisms in soilless cultures. Agron Sustain Dev 31:191–203. https://doi.org/10.1051/agro/2010018
Sutton JC, Sopher CR, Owen-Going TN et al (2006) Etiology and epidemiology of Pythium root rot in hydroponic crops: current knowledge and perspectives. Summa Phytopathol 32:307–321. https://doi.org/10.1590/S0100-54052006000400001
Stanghellini ME, Rasmussen SL (1994) Hydroponics: a solution for zoosporic pathogens. Plant Dis 78:1129–1138. https://doi.org/10.1094/PD-78-1129
Alhussaen K (2006) Pythium and Phytophthora associated with root disease of hydroponic lettuce. PhD thesis, University of Technologie Sydney. https://opus.lib.uts.edu.au/handle/10453/36864
Rakocy JE, Maseer PM, M. Losordo T (2006) Recirculating aquaculture tank production systems – integrated fish and plant culture. South Reg Aquac Cent. https://www.semanticscholar.org/paper/Recirculating-Aquaculture-Tank-Production-Systems-%3A-Rakocy-Masser/7db4a54e7ffab14830af2551e84d87959bd230d4#citing-papers
Bittsanszky A, Gyulai G, Junge R, et al (2015) Plant protection in ecocycle-based agricultural systems: aquaponics as an example. In: International Plant Protection Congress (IPPC). Berlin, Germany, pp 2–3. https://www.researchgate.net/publication/281714199_Plant_protection_in_ecocyclebased_agricultural_systems_aquaponics_as_an_example
Nemethy S, Bittsanszky A, Schmautz Z et al (2016) Protecting plants from pests and diseases in aquaponic systems. Ecological Footprint in Central Europe. The University College of Tourism and Ecology Press, Sucha Beskidzka, Poland, pp 1–8
Folorunso EA, Roy K, Gebauer R, et al (2020) Integrated pest and disease management in aquaponics: a metadata-based review. Rev Aquac 13:971–995. https://doi.org/10.1111/raq.12508
Stouvenakers G, Dapprich P, Massart S, Jijakli MH (2019) Ch 14: Plant pathogens and control strategies in aquaponics. In: Joyce A, Kotzen B, Burnell GM (eds) Simon Goddek. Aquaponics food production systems. Springer, Cham, pp 353–378
Paulitz TC (1997) Biological control of root pathogens in soilless and hydroponic systems. HortScience 32:193–196
Postma J, van Os E, Bonants PJM (2008) Ch 10 – Pathogen detection and management strategies in soilless plant growing system. In: Soilless culture: theory and practice, third edition. Elsevier B.V., pp 425–457. https://www.sciencedirect.com/science/article/pii/B9780444529756500125?via%3Dihub
Montagne V, Capiaux H, Barret M et al (2017) Bacterial and fungal communities vary with the type of organic substrate: implications for biocontrol of soilless crops. Environ Chem Lett 15:537–545. https://doi.org/10.1007/s10311-017-0628-0
Deacon JW, Entwistle AR, Deacon JW et al (1988) Biocontrol of soil-borne plant pathogens with introduced inocula [and discussion]. Philos Trans R Soc Lond B Biol Sci 318:249–264
Stouvenakers G, Massart S, Depireux P, Jijakli MH (2020) Microbial origin of aquaponic water suppressiveness against Pythium aphanidermatum lettuce root rot disease. Microorganisms 8:1–25. https://doi.org/10.3390/microorganisms8111683
Schlatter D, Kinkel L, Thomashow L et al (2017) Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107:1284–1297. https://doi.org/10.1094/PHYTO-03-17-0111-RVW
Expósito RG, de Bruijn I, Postma J, Raaijmakers JM (2017) Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front Microbiol 8:1–12. https://doi.org/10.3389/fmicb.2017.02529
Niem JM, Billones-Baaijens R, Stodart B, Savocchia S (2020) Diversity profiling of grapevine microbial endosphere and antagonistic potential of endophytic Pseudomonas against grapevine trunk diseases. Front Microbiol 11:1–19. https://doi.org/10.3389/fmicb.2020.00477
Liao H, Huang L, Li N et al (2021) Auxiliary rapid identification of pathogenic and antagonistic microorganisms associated with Coptis chinensis root rot by high-throughput sequencing. Sci Rep 11:1–15. https://doi.org/10.1038/s41598-021-90489-9
Balouiri M, Sadiki M, Ibnsouda SK (2016) Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal 6:71–79. https://doi.org/10.1016/j.jpha.2015.11.005
Raymaekers K, Ponet L, Holtappels D et al (2020) Screening for novel biocontrol agents applicable in plant disease management – a review. Biol Control 144:104240. https://doi.org/10.1016/j.biocontrol.2020.104240
Eck M, Szekely I, Massart S, Jijakli MH (2021) Ecological study of aquaponics bacterial microbiota over the course of a lettuce growth cycle. Water 13:21
Khan A, Sutton JC, Grodzinski B (2003) Effects of Pseudomonas chlororaphis on Pythium aphanidermatum and root rot in peppers grown in small-scale hydroponic troughs. Biocontrol Sci Technol 13:615–630. https://doi.org/10.1080/0958315031000151783
Chatterton S, Sutton JC, Boland GJ (2004) Timing Pseudomonas chlororaphis applications to control Pythium aphanidermatum, Pythium dissotocum, and root rot in hydroponic peppers. Biol Control 30:360–373. https://doi.org/10.1016/j.biocontrol.2003.11.001
Liu W, Sutton JC, Grodzinski B et al (2007) Biological control of Pythium root rot of chrysanthemum in small-scale hydroponic units. Phytoparasitica 35:159–178. https://doi.org/10.1007/BF02981111
Eck M, Sare AR, Massart S et al (2019) Exploring bacterial communities in aquaponic systems. Water 11:1–16. https://doi.org/10.3390/w11020260
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877. https://doi.org/10.1101/gr.9.9.868
Jansson H-B, Thiman L (1992) A preliminary study of chemotaxis of zoospores of the nematode-parasitic fungus Catenaria anguillulae. Mycologia 84:109. https://doi.org/10.2307/3760409
Reddy PP (2014) Plant growth promoting rhizobacteria for horticultural crop protection, 1st editio. Springer India. https://link.springer.com/content/pdf/10.1007/978-81-322-1973-6.pdf
Sirakov I, Lutz M, Graber A et al (2016) Potential for combined biocontrol activity against fungal fish and plant pathogens by bacterial isolates from a model aquaponic system. Water 8:1–7. https://doi.org/10.3390/w8110518
Wu Q, Dou X, Wang Q et al (2018) Isolation of β-1,3-glucanase-producing microorganisms from Poria cocos cultivation soil via molecular biology. Molecules 23:1555. https://doi.org/10.3390/molecules23071555
Davis KER, Joseph SJ, Janssen PH (2005) Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol 71:826–834. https://doi.org/10.1128/AEM.71.2.826-834.2005
El-Tarabily KA, Sivasithamparam K (2006) Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol Biochem 38:1505–1520. https://doi.org/10.1016/j.soilbio.2005.12.017
Williams ST, Wellington EMH (1982) Principles and problems of selective isolation of microbes. In: Bu’lock JD (ed) Bioactive metabolic products: search and discovery. Academic Press, London, pp 9–26. https://catalog.loc.gov/vwebv/search?searchCode=LCCN&searchArg=81068969&searchType=1&permalink=y
El-Tarabily KA (2006) Rhizosphere-competent isolates of streptomycete and non-streptomycete actinomycetes capable of producing cell-wall-degrading enzymes to control Pythium aphanidermatum damping-off disease of cucumber. Can J Bot 84:211–222. https://doi.org/10.1139/B05-153
Evangelista-Martínez Z (2014) Isolation and characterization of soil Streptomyces species as potential biological control agents against fungal plant pathogens. World J Microbiol Biotechnol 30:1639–1647. https://doi.org/10.1007/s11274-013-1568-x
Nadeem SM, Naveed M, Ayyub M, Khan MY (2016) Potential, limitations and future prospects of Pseudomonas spp. for sustainable agriculture and environment: a review. Soil Environ 35:106–145
Thongkamngam T, Jaenaksorn T (2017) Fusarium oxysporum (F221-B) as biocontrol agent against plant pathogenic fungi in vitro and in hydroponics. Plant Prot Sci 53:85–95. https://doi.org/10.17221/59/2016-PPS
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511. https://doi.org/10.1093/jexbot/52.suppl_1.487
Boers SA, Jansen R, Hays JP (2019) Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur J Clin Microbiol Infect Dis 38:1059–1070
Stefani FOP, Bell TH, Marchand C et al (2015) Culture-dependent and -independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PLoS ONE 10:1–16. https://doi.org/10.1371/journal.pone.0128272
Forbes JD, Knox NC, Ronholm J et al (2017) Metagenomics: the next culture-independent game changer. Front Microbiol 8:1–21. https://doi.org/10.3389/fmicb.2017.01069
Garbeva P, Van Veen JA, Van Elsas JD (2004) Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270. https://doi.org/10.1146/annurev.phyto.42.012604.135455
Torsvik V, Sørheim R, Goksøyr J (1996) Total bacterial diversity in soil and sediment communities – a review. J Ind Microbiol Biotechnol 17:170–178. https://doi.org/10.1007/bf01574690
Köhl J, Kolnaar R, Ravensberg WJ (2019) Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front Plant Sci 10:1–19. https://doi.org/10.3389/fpls.2019.00845
Martin FN, Loper JE (1999) Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. CRC Crit Rev Plant Sci 18:111–181
Adams PB (1971) Pythium aphanidermatum oospore germination as affected by time, temperature, and pH. Phytopathology 61:1149–1150
Song D, Chen X, Xu M et al (2019) Adaptive evolution of Sphingobium hydrophobicum C1T in electronic waste contaminated river sediment. Front Microbiol 10:1–16. https://doi.org/10.3389/fmicb.2019.02263
Stolz A, Schmidt-Maag C, Denner EBM et al (2000) Description of Sphingomonas xenophaga sp. nov. for strains BN6(T) and N, N which degrade xenobiotic aromatic compounds. Int J Syst Evol Microbiol 50:35–41. https://doi.org/10.1099/00207713-50-1-35
Cardinale M, Grube M, Erlacher A et al (2015) Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ Microbiol 17:239–252. https://doi.org/10.1111/1462-2920.12686
Schmautz Z, Graber A, Jaenicke S, et al (2017) Microbial diversity in different compartments of an aquaponics system. Arch Microbiol 199:613–620. https://doi.org/10.1007/s00203-016-1334-1
Sare AR, Stouvenakers G, Eck M et al (2020) Standardization of plant microbiome studies: which proportion of the microbiota is really harvested? Microorganisms 8:17. https://doi.org/10.3390/microorganisms8030342
Ortega RA, Mahnert A, Berg C et al (2016) The plant is crucial: specific composition and function of the phyllosphere microbiome of indoor ornamentals. FEMS Microbiol Ecol 92:12. https://doi.org/10.1093/femsec/fiw173
Wanees AE, Zaslow SJ, Potter SJ et al (2018) Draft genome sequence of the plant growth-promoting Sphingobium sp strain aew4, isolated from the rhizosphere of the beachgrass Ammophila breviligulata. Genome Announc 6:2. https://doi.org/10.1128/genomeA.00410-18
Miller CD, Hall K, Liang YN et al (2004) Isolation and characterization of polycyclic aromatic hydrocarbon-degrading mycobacterium isolates from soil. Microb Ecol 48:230–238. https://doi.org/10.1007/s00248-003-1044-5
Bisht S, Pandey P, Bhargava B et al (2015) Bioremediation of polyaromatic hydrocarbons (PAHs) using rhizosphere technology. Brazilian J Microbiol 46:7–21. https://doi.org/10.1590/S1517-838246120131354
Kyselková M, Kopeck J, Frapolli M et al (2009) Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease. ISME J 3:1127–1138. https://doi.org/10.1038/ismej.2009.61
Burgos-Garay ML, Hong C, Moorman GW (2014) Interactions of heterotrophic bacteria from recycled greenhouse irrigation water with plant pathogenic Pythium. HortScience 49:961–967. https://doi.org/10.21273/hortsci.49.7.961
Sanchez FA, Vivian-Rogers VR, Urakawa R (2019) Tilapia recirculating aquaculture systems as a source of plant growth promoting bacteria. Aquac Res 50:2054–2065. https://doi.org/10.1111/are.14072
Fattah AM, Sayed E (2006) Tilapia culture, CABI Publi. https://www.aquacultureinafrica.com/?p=1623
Faria S, Joao I, Jordao L (2015) General overview on nontuberculous Mycobacteria, biofilms, and human Infection. J Pathog 2015:1–10. https://doi.org/10.1155/2015/809014
Alabouvette C, Cordier C (2011) Risks of microbial biocontrol agents and regulation: are they in balance? In: Ehlers R-U (ed) Regulation of Biological Control Agents. Springer, Dordrecht, pp 1–416
Von Graevenitz A, Weinstein J (1971) Pathogenic significance of Pseudomonas fluorescens and Pseudomonas putida. Yale J Biol Med 44:265–273
Amaike S, Keller NP (2011) Aspergillus flavus. Annu Rev Phytopathol 49:107–133. https://doi.org/10.1146/annurev-phyto-072910-095221
Khan R, Ghazali FM, Mahyudin NA, Samsudin NIP (2021) Biocontrol of aflatoxins using non-aflatoxigenic Aspergillus flavus: A literature review. J Fungi 7:13. https://doi.org/10.3390/jof7050381
Shanmugan V, Sakurana Varma A (1999) Effect of native antagonists against Pythium aphanidermatum, the causal organism of rhizome rot in ginger. J Mycol plant Pathol 29:375–379
Acknowledgements
The authors are grateful to the laboratory staff and their supervisors for their valuable assistance and reviewing. We especially thank the trainee Fanny Vautrin who helped screen the biocontrol agents.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by GS. The first draft of the manuscript was written by GS, and all authors commented on the previous versions of the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Stouvenakers, G., Massart, S. & Jijakli, M.H. First Study Case of Microbial Biocontrol Agents Isolated from Aquaponics Through the Mining of High-Throughput Sequencing Data to Control Pythium aphanidermatum on Lettuce. Microb Ecol 86, 1107–1119 (2023). https://doi.org/10.1007/s00248-022-02126-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02126-1