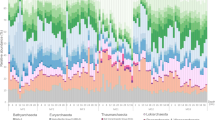
Abstract
Bathyarchaeota are believed to have roles in the carbon cycle in marine systems. However, the ecological knowledge of Bathyarchaeota is limited in peatland ecosystems. Here, we investigated the vertical distribution of Bathyarchaeota community structure using quantitative PCR and high-throughput sequencing technology of ribosomal 16S rRNA gene integrated with detailed chemical profiling in the Dajiuhu Peatland, central China. Eight archaeal phyla were observed in peat samples, which mainly composed of Bathyarchaeota with a mean relative abundance about 88%, followed by Thaumarchaeota (9%). Bathyarchaeota were further split into 17 subgroups, and some subgroups showed habitat specificity to peat horizons with distinct lithological and physicochemical properties, for example, Bathy-6 and Bathy-15 had preference for the acrotelm, Bathy-5b, Bathy-16, and Bathy-19 were enriched in the catotelm, Bathy-5a, Bathy-8, and Bathy-11 were specific for the clay horizon. This spatial distribution pattern of archaeal communities along peat profile was mainly influenced by water content as indicated by RDA ordination and permutational MANOVA, whereas organic matter content exclusively affected Bathyarchaeota distribution along the peat profile significantly. The abundance of archaeal 16S rRNA genes ranged from 105 to 107 copies per gram dry sediment, and the highest archaeal biomass was observed in the periodically oxic mesotelm horizon with more dynamic archaeal interaction relationship as indicated by the network analysis. Bathyarchaeota dominated the archaeal interaction network with 82% nodes, 96% edges, and 71% keystone species. Our results provide an overview of the archaeal population, community structure, and relationship with environmental factors that affect the vertical distribution of archaeal communities and emphasize the ecology of bathyarchaeotal lineages in terrestrial peatland ecosystems.



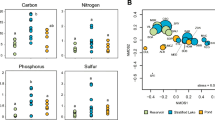
source of variation in Aithison distance. In b, color parts denoted significant factors at 0.05 level for explanation of variance resource, and unknown part represented the residuals from permutational MANOVA analysis




Similar content being viewed by others
Data Availability
Sequence data obtained from this study were deposited in the National Omics Data Encyclopedia (NODE) under the accession number of OEP001037.
Code Availability
Not applicable.
References
Yu Z, Loisel J, Brosseau DP, Beilman DW, Hunt SJ (2010) Global peatland dynamics since the last glacial maximum. Geophys Res Lett 37:L13402. https://doi.org/10.1029/2010GL043584
Limpens J, Heijmans MMPD, Berendse F (2006) The nitrogen cycle in boreal peatlands. In: Wieder RK, Vitt DH (eds) Boreal peatland ecosystems. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 195–230
Menon S, Denman KL, Brasseur G, Chidthaisong A, Ciais P, Cox PM, Dickinson RE, Hauglustaine D, Heinze C, Holland E, Jacob D, Lohmann U, Ramachandran S, da Silva L, Dias P, Wofsy SC, Zhang X (2007) Couplings between changes in the climate system and biogeochemistry. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL (eds) Contribution of Working Group Ito the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kindom, pp 499–587
Tveit A, Schwacke R, Svenning MM, Urich T (2013) Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J 7:299–311. https://doi.org/10.1038/ismej.2012.99
Cadillo-Quiroz H, Bräuer S, Yashiro E, Sun C, Yavitt J, Zinder S (2006) Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ Microbiol 8:1428–1440. https://doi.org/10.1111/j.1462-2920.2006.01036.x
Akiyama M, Shimizu S, Sakai T, Ioka S, Ishijima Y, Naganuma T (2011) Spatiotemporal variations in the abundances of the prokaryotic rRNA genes, pmoA, and mcrA in the deep layers of a peat bog in Sarobetsu-genya wetland, Japan. Limnology 12:1–9. https://doi.org/10.1007/s10201-010-0315-3
Zhou L, Liu X, Dong X (2014) Methanospirillum psychrodurum sp. nov., isolated from wetland soil. Int J Syst Evol Microbiol 64:638–641. https://doi.org/10.1099/ijs.0.057299-0
Kotsyurbenko O, Friedrich M, Simankova M, Nozhevnikova A, Golyshin P, Timmis K, Conrad R (2007) Shift from acetoclastic to H2-dependent methanogenesis in a West Siberian peat bog at low pH values and isolation of an acidophilic Methanobacterium strain. Appl Environ Microbiol 73:2344–2348. https://doi.org/10.1128/AEM.02413-06
Cadillo-Quiroz H, Bräuer SL, Goodson N, Yavitt JB, Zinder SH (2014) Methanobacterium paludis sp. nov. and a novel strain of Methanobacterium lacus isolated from northern peatlands. Int J Syst Evol Microbiol 64:1473–1480. https://doi.org/10.1099/ijs.0.059964-0
Löscher C, Kock A, Koenneke M, LaRoche J, Bange HW, Schmitz RA (2012) Production of oceanic nitrous oxide by ammonia-oxidizing archaea. Biogeosciences 9:2419–2429. https://doi.org/10.5194/bg-9-2419-2012
Siljanen HM, Alves RJ, Ronkainen JG, Lamprecht RE, Bhattarai HR, Bagnoud A, Marushchak ME, Martikainen PJ, Schleper C, Biasi C (2019) Archaeal nitrification is a key driver of high nitrous oxide emissions from arctic peatlands. Soil Biol Biochem 137:107539. https://doi.org/10.1016/j.soilbio.2019.107539
Xiang X, Wang R, Wang H, Gong L, Man B, Xu Y (2017) Distribution of Bathyarchaeota communities across different terrestrial settings and their potential ecological functions. Sci Rep 7:45028. https://doi.org/10.1038/srep45028
Bai Y, Wang J, Zhan Z, Guan L, Jin L, Zheng G, Huang Z (2018) The variation of microbial communities in a depth profile of peat in the Gahai Lake Wetland Natural Conservation area. Geomicrobiol J 35:484–490. https://doi.org/10.1080/01490451.2017.1392651
Wen X, Unger V, Jurasinski G, Koebsch F, Horn F, Rehder G, Sachs T, Zak D, Lischeid G, Knorr K-H (2018) Predominance of methanogens over methanotrophs in rewetted fens characterized by high methane emissions. Biogeosciences 15:6519–6536. https://doi.org/10.5194/bg-15-6519-2018
Meng J, Xu J, Qin D, He Y, Xiao X, Wang F (2014) Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J 8:650–659. https://doi.org/10.1038/ismej.2013.174
Yu T, Wu W, Liang W, Lever MA, Hinrichs K-U, Wang F (2018) Growth of sedimentary Bathyarchaeota on lignin as an energy source. PNAS 115:6022–6027. https://doi.org/10.1073/pnas.1718854115
Schellekens J, Buurman P, Kuyper TW (2012) Source and transformations of lignin in Carex-dominated peat. Soil Biol Biochem 53:32–42. https://doi.org/10.1016/j.soilbio.2012.04.030
Chen Y, Li S, Yu Z, Chen Y, Mi T, Zhen Y (2020) Characteristics of the Bathyarchaeota community in surface sediments from the southern Yellow Sea and northern East China sea. Estuarine, Coastal Shelf Sci 235:106595. https://doi.org/10.1016/j.ecss.2020.106595
Fillol M, Sànchez-Melsió A, Gich F, Borrego MC (2015) Diversity of Miscellaneous Crenarchaeotic Group archaea in freshwater karstic lakes and their segregation between planktonic and sediment habitats. FEMS Microbiol Ecol 91:fiv020. https://doi.org/10.1093/femsec/fiv020
Pan J, Zhou Z, Béjà O, Cai M, Yang Y, Liu Y, Gu JD, Li M (2020) Genomic and transcriptomic evidence of light-sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome 8:43. https://doi.org/10.1186/s40168-020-00820-1
Zou D, Pan J, Liu Z, Zhang C, Liu H, Li M (2020) The distribution of Bathyarchaeota in surface sediments of the Pearl River estuary along salinity gradient. Front Microbiol 11:285. https://doi.org/10.3389/fmicb.2020.00285
He Y, Li M, Perumal V, Feng X, Fang J, Xie J, Sievert SM, Wang F (2016) Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol 1:16035. https://doi.org/10.1038/NMICROBIOL.2016.35
Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs KU, Teske AP (2015) Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol 18:1200–1211. https://doi.org/10.1111/1462-2920.13142
Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW (2015) Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. https://doi.org/10.1126/science.aac7745
Lloyd K, Schreiber L, Petersen D, Kjeldsen K, Lever M, Steen A, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen B (2013) Predominant Archaea in marine sediments degrade detrital proteins. Nature 496:215–218. https://doi.org/10.1038/nature12033
Bräuer LS, Basiliko N, Siljanen MPH, Zinder HS (2020) Methanogenic archaea in peatlands. FEMS Microbiol Lett 367:fnaa172. https://doi.org/10.1093/femsle/fnaa172
Zhou Z, Pan J, Wang F, Gu J, Li M (2018) Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol Rev 42:639–655. https://doi.org/10.1093/femsre/fuy023
Zhou Z, Zhang G-X, Xu Y-B, Gu J-D (2018) Successive transitory distribution of Thaumarchaeota and partitioned distribution of Bathyarchaeota from the Pearl River estuary to the northern South China Sea. Appl Microbiol Biotechnol 102:8035–8048. https://doi.org/10.1007/s00253-018-9147-6
Fillol M, Auguet JC, Casamayor EO, Borrego CM (2016) Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage. ISME J 10:665–677. https://doi.org/10.1038/ismej.2015.143
Xiang X, Wang H, Gong L, Liu Q (2014) Vertical variations and associated ecological function of bacterial communities from Sphagnum to underlying sediments in Dajiuhu Peatland. Sci China: Earth Sci 57:1013–1020. https://doi.org/10.1007/s11430-013-4752-9
Xiang X, Wang H, Tian W, Wang R, Gong L, Xu Y, Man B (2020) Composition and function of bacterial communities of bryophytes and their underlying sediments in the Dajiuhu Peatland, central China. J Earth Sci. https://doi.org/10.1007/s12583-020-1391-x
Coolen MJ, Hopmans EC, Rijpstra WIC, Muyzer G, Schouten S, Volkman JK, Damsté JSS (2004) Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: response of methanogens and methanotrophs to environmental change. Org Geochem 35:1151–1167. https://doi.org/10.1016/j.orggeochem.2004.06.009
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072. https://doi.org/10.1128/AEM.00595-10
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67:4399–4406. https://doi.org/10.1128/aem.67.10.4399-4406.2001
Chao A, Lee s-M (1992) Estimating the number of classes via sample coverage. J Am Stat Assoc 87:210–217. https://doi.org/10.1080/01621459.1992.10475194
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. https://doi.org/10.1016/0006-3207(92)91201-3
George D, Mallery P (2003) SPSS for Windows Step by Step: a simple Guide and Reference, 11.0 update, 4th Edn. Allyn & Bacon, Boston
Mei W, Yu G, Lai J, Rao Q, Umezawa Y (2018) basicTrendline: Add trendline and confidence interval of basic regression models to plot. R package version 2.0.3. https://cran.r-project.org/web/packages/basicTrendline
Revelle W, Revelle MW (2015) psych: Procedures for personality and psychological research. R package version 2.1.9. https://cran.r-project.org/web/packages/psych
Rohart F, Gautier B, Singh A, Lê Cao K-A (2017) mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 13:e1005752
Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M (2007) The vegan package. R package version 2.5-7. https://cran.r-project.org/web/packages/vegan
Braak CJFt, Smilauer P (2012) Canoco reference manual and user’s guide: software for ordination, version 5.0. Microcomputer Power, Ithaca
Friedman J, Alm EJ (2012) Inferring correlation networks from genomic survey data. PLoS Comput Biol 8:e1002687. https://doi.org/10.1371/journal.pcbi.1002687
Csardi G, Nepusz T (2006) The igraph software package for complex network research. R package version 1.2.6. https://cran.r-project.org/web/packages/igraph
Berry D, Widder S (2014) Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5:219. https://doi.org/10.3389/fmicb.2014.00219
Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, Brookes P, Gilbert J, xu J (2015) Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J 10:1891–1901. https://doi.org/10.1038/ismej.2015.261
Pala C, Molari M, Nizzoli D, Bartoli M, Viaroli P, Manini E (2018) Environmental drivers controlling bacterial and archaeal abundance in the sediments of a Mediterranean lagoon ecosystem. Curr Microbiol 75:1147–1155. https://doi.org/10.1007/s00284-018-1503-3
Chen X, Andersen TJ, Morono Y, Inagaki F, Jørgensen BB, Lever MA (2017) Bioturbation as a key driver behind the dominance of Bacteria over Archaea in near-surface sediment. Sci Rep 7:2400. https://doi.org/10.1038/s41598-017-02295-x
Zhang J, Yang Y, Zhao L, Li Y, Xie S, Liu Y (2014) Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl Microbiol Biotechnol 99:3291–3302. https://doi.org/10.1007/s00253-014-6262-x
Metje M, Frenzel P (2005) Effect of temperature on anaerobic ethanol oxidation and methanogenesis in acidic peat from a northern wetland. Appl Environ Microbiol 71:8191–8200. https://doi.org/10.1128/aem.71.12.8191-8200.2005
Jungbluth S, Amend J, Rappe M (2016) Metagenome sequencing and 98 microbial genomes from Juan de Fuca Ridge flank subsurface fluids. Sci Data 4:170037. https://doi.org/10.7287/PEERJ.PREPRINTS.2613V1
Jay Z, Beam J, Dlakić M, Rusch D, Kozubal M, Inskeep W (2018) Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nat Microbiol 3:732–740. https://doi.org/10.1038/s41564-018-0163-1
Wang Y, Wegener G, Hou J, Wang F, Xiao X (2019) Expanding anaerobic alkane metabolism in the domain of Archaea. Nat Microbiol 4:595–602. https://doi.org/10.1038/s41564-019-0364-2
Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, Tyson GW (2016) Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol 1:16170. https://doi.org/10.1038/nmicrobiol.2016.170
Pester M, Schleper C, Wagner M (2011) The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306. https://doi.org/10.1016/j.mib.2011.04.007
Niu M, Zhou F, Yang Y, Sun Y, Zhu T, Shen F (2021) Abundance and composition of airborne archaea during springtime mixed dust and haze periods in Beijing. China. Sci Total Environ 752:141641. https://doi.org/10.1016/j.scitotenv.2020.141641
Lin X, Handley KM, Gilbert JA, Kostka JE (2015) Metabolic potential of fatty acid oxidation and anaerobic respiration by abundant members of Thaumarchaeota and Thermoplasmata in deep anoxic peat. ISME J 9:2740–2744. https://doi.org/10.1038/ismej.2015.77
Wang D (2018) Characteristics and influence factors of temporal and spatial variation of dissolved organic carbon in pore water from the Dajiuhu peatland,Hubei province. Master's Thesis, China University of Geosciences (Wuhan)
Mwirichia R, Alam I, Rashid M, Vinu M, Ba-Alawi W, Anthony Kamau A, Kamanda Ngugi D, Göker M, Klenk H-P, Bajic V, Stingl U (2016) Metabolic traits of an uncultured archaeal lineage -MSBL1- from brine pools of the Red Sea. Sci Rep 6:19181. https://doi.org/10.1038/srep19181
Guy L, Ettema TJ (2011) The archaeal ‘TACK’superphylum and the origin of eukaryotes. Trends Microbiol 19:580–587. https://doi.org/10.1016/j.tim.2011.09.002
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nat Environ Pollut Technol 521:173–179. https://doi.org/10.1038/nature14447
Etto R, Cruz L, Jesus E, Galvão C, Galvão F, Souza E, Pedrosa F, Steffens M (2012) Prokaryotic communities of acidic peatlands from the southern Brazilian Atlantic Forest. Braz J Microbiol 43:661–674. https://doi.org/10.1590/S1517-83822012000200031
Fan X, Xing P (2016) Differences in the composition of archaeal communities in sediments from contrasting zones of lake Taihu. Front Microbiol 7:1510. https://doi.org/10.3389/fmicb.2016.01510
Navarro-Noya YE, Valenzuela-Encinas C, Sandoval-Yuriar A, Jiménez-Bueno NG, Marsch R, Dendooven L (2015) Archaeal communities in a heterogeneous hypersaline-alkaline soil. Archaea 2015:646820. https://doi.org/10.1155/2015/646820
Wang S, Dong RM, Dong CZ, Huang L, Jiang H, Wei Y, Feng L, Liu D, Yang G, Zhang C, Dong H (2012) Diversity of microbial plankton across the Three Gorges Dam of the Yangtze River, China. Geosci Front 3:335–349. https://doi.org/10.1016/j.gsf.2011.11.013
Hawkins A, Johnson K, Bräuer S (2014) Southern Appalachian peatlands support high archaeal diversity. Microb Ecol 67:587–602. https://doi.org/10.1007/s00248-013-0352-7
Liu X, Li M, Castelle CJ, Probst AJ, Zhou Z, Pan J, Liu Y, Banfield JF, Gu J-D (2018) Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome 6:102. https://doi.org/10.1186/s40168-018-0488-2
Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM (2015) Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25:690–701. https://doi.org/10.1016/j.cub.2015.01.014
Genderjahn S, Alawi M, Mangelsdorf K, Horn F, Wagner D (2018) Desiccation- and saline-tolerant bacteria and archaea in Kalahari Pan sediments. Front Microbiol 9:2082. https://doi.org/10.3389/fmicb.2018.02082
Angel R, Soares MIM, Ungar ED, Gillor O (2010) Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J 4:553–563. https://doi.org/10.1038/ismej.2009.136
Andersen R, Chapman SJ, Artz RRE (2013) Microbial communities in natural and disturbed peatlands: A review. Soil Biol Biochem 57:979–994. https://doi.org/10.1016/j.soilbio.2012.10.003
Holden J (2006) Chapter 14 Peatland hydrology. In: Martini IP, Martínez Cortizas A, Chesworth W (eds) Developments in earth surface processes. Elsevier, pp 319–346
Juottonen H, Fontaine L, Wurzbacher C, Drakare S, Peura S, Eiler A (2020) Archaea in boreal Swedish lakes are diverse, dominated by Woesearchaeota and follow deterministic community assembly. Environ Microbiol 22:3158–3171. https://doi.org/10.1111/1462-2920.15058
Putkinen A, Juottonen H, Juutinen S, Tuittila E-S, Fritze H, Yrjälä K (2009) Archaeal rRNA diversity and methane production in deep boreal peat. FEMS Microbiol Ecol 70:87–98. https://doi.org/10.1111/j.1574-6941.2009.00738.x
Huber H, Stetter K (2015) Desulfurococcales ord. nov. In: Whitman, WB, Rainey, F, Kämpfer, P, Trujillo, M, Chun, J, DeVos, P, Hedlund, B, Dedysh, S (eds) In Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley, in association with Bergey’s Manual Trust, pp 1–2
Nkamga V, Drancourt M (2016) Methanomassiliicoccales. In: Whitman, WB, Rainey, F, Kämpfer, P, Trujillo, M, Chun, J, DeVos, P, Hedlund, B, Dedysh, S (eds) In Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley, in association with Bergey’s Manual Trust, pp 1–2
Chen L, Hu M, Huang L, Hua Z, Kuang J, Li S, Shu W (2015) Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J 9:1579–1592. https://doi.org/10.1038/ismej.2014.245
Foster JW (2004) Escherichia coli acid resistance: Tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. https://doi.org/10.1038/nrmicro1021
Guazzaroni M-E, Morgante V, Mirete S, Gonzalez-Pastor E (2012) Novel acid resistance genes from the metagenome of the Tinto River, an extremely acidic environment. Environ Microbiol 15:1088–1102. https://doi.org/10.1111/1462-2920.12021
Xiang X (2019) Environmental genomics study of archaea in Dajiuhu Peatland. Doctoral Thesis, China University of Geosciences (Wuhan)
Cools N, Vesterdal L, De Vos B, Vanguelova E, Hansen K (2014) Tree species is the major factor explaining C: N ratios in European forest soils. For Ecol Manage 311:3–16. https://doi.org/10.1016/j.foreco.2013.06.047
Shin J-H, An N-H, Lee S-M, Ok J-H, Lee B-W (2016) Estimation of N mineralization potential and N mineralization rate of organic amendments as affected by C: N ratio and temperature in paddy soil. Korean J Soil Sci Fert 49:712–719. https://doi.org/10.7745/KJSSF.2016.49.6.712
Fan K, Cardona C, Li Y, Shi Y, Xiang X, Shen C, Wang H, Gilbert JA, Chu H (2017) Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol Biochem 113:275–284. https://doi.org/10.1016/j.soilbio.2017.06.020
Jones CM, Hallin S (2019) Geospatial variation in co-occurrence networks of nitrifying microbial guilds. Mol Ecol 28:293–306. https://doi.org/10.1111/mec.14893
Newman MEJ (2003) The structure and function of complex networks. SIAM Rev 45:167–256. https://doi.org/10.1016/S0010-4655(02)00201-1
Trosvik P, de Muinck EJ (2015) Ecology of bacteria in the human gastrointestinal tract—identification of keystone and foundation taxa. Microbiome 3:44. https://doi.org/10.1186/s40168-015-0107-4
Acknowledgements
We thank Miss Renju Liu and Mr. Jiang Liu (Third Institute of Oceanography), Ziqi Zhang (Shennongjia Nature Reserve), and Jinjiang Pan (China University of Geosciences (Wuhan)) for their assist in sampling and experience. We also thank Dr. Chuanlun Zhang (Southern University of Science and Technology), Meng Li (Shenzhen University), Olli H Tuovinen (Ohio State University), the editor, and the anonymous reviewers for their comments on an early version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (No. 41572325), the Fundamental Research Funds for the Central Universities, China University of Geosciences (Wuhan) (CUGCJ1703, CUGQY1922), and Open Research Fund of Hubei Key Laboratory of Critical Zone Evolution, China University of Geosciences, Wuhan, China (2021F10).
Author information
Authors and Affiliations
Contributions
X.X. and H.W. conceived and designed this experiment; X.X., Y.X., and W.T. performed the experiment and collected data; X.X., B.M., and L.G. analyzed those data. X.X., H.W., and H. Y. discussed the results and revised the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiang, X., Wang, H., Man, B. et al. Diverse Bathyarchaeotal Lineages Dominate Archaeal Communities in the Acidic Dajiuhu Peatland, Central China. Microb Ecol 85, 557–571 (2023). https://doi.org/10.1007/s00248-022-01990-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-01990-1