Abstract
Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli cause severe health hazards. Migratory birds are reservoirs and transmitters of many pathogens including ESBL-producing E. coli. To examine migratory birds as potential carriers of ESBL-producing E. coli and E. coli-carrying antibiotic resistance genes, 55 PCR-positive E. coli isolates were screened using the disk diffusion method, double-disk synergy test, and further polymerase chain reaction (PCR) tests. Genes encoding resistance to tetracycline [tetA, 100% (35/35); tetB, 31.43% (11/35)], fluoroquinolone [qnrA, 35.71% (10/28); qnrB, 25% (7/28)], and streptomycin [aadA1, 90.24% (37/41)] were detected in the isolated E. coli. Of the 55 E. coli isolates, 21 (38.18%) were ESBL producers, and all of them were multidrug resistant. All the ESBL-producing E. coli isolates harbored at least two or more beta-lactamase genes, of which blaTEM, blaCMY, blaCTX-M, and blaSHV were detected in 95.24%, 90.48%, 85.71%, and 42.86% of isolates, respectively. All the beta-lactamase genes were present in four of the ESBL-producing E. coli isolates. Furthermore, 95.24% of ESBL-producing E. coli isolates were positive for one or more antibiotic resistance genes. To the best of our knowledge, this is the first study to detect E. coli-carrying antibiotic resistance genes including beta-lactamase blaCMY and blaSHV originating from migratory birds in Bangladesh. These results suggest that migratory birds are potential carriers of ESBL-producing E. coli along with other clinically important antibiotic resistance genes which may have detrimental impacts on human health.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) is a major global health concern of the twenty-first century affecting all the components of health including animals, humans, and environments [1]. Selective pressure resulting from the overuse of antibiotics is a major driver for the development of resistance [2]. Migratory birds during the course of their travels acquire and transmit antibiotic-resistant bacteria and/or AMR genes across environments [3, 4].
Escherichia coli, commensal bacteria, are regarded as excellent indicative species to reveal the transmission and dissemination of AMR via water contaminated by fecal materials [5, 6]. In addition, E. coli are abundant in different water bodies where migratory birds reside and are involved in retaining AMR in those locations [7]. Thus, waterfowl including migratory birds have the potential to be sentinels of AMR E. coli in the environment.
Cephalosporins comprise a widely used group of beta-lactam antibiotics in humans. The dissemination of extended-spectrum beta-lactamase (ESBL)-producing E. coli in humans and animals has increasingly emerged around the globe. E. coli increasingly encounters beta-lactam antibiotics and has developed resistance, causing common community-related infections [8]. The production of beta-lactamase, a degradative enzyme, usually mediates beta-lactam resistance in E. coli pathogens [9]. The existence of ESBL resistance within E. coli has emerged as a global crisis in antibiotic choices [10]. Different classes of antimicrobials, e.g., tetracyclines, aminoglycosides, fluoroquinolones, and trimethoprim–sulfamethoxazole, have been commonly become ineffective in beta-lactamase-producing E. coli resulting in increased morbidity–mortality, prolonging hospitalization, increased treatment cost, and deterioration of healthcare systems [10, 11]. Two plasmid-borne classes of beta-lactam enzyme, ESBL and AmpC beta-lactamases (AmpC), impact the health sector negatively throughout the world, causing resistance to beta-lactam antibiotics [12]. Additionally, CTX-M, TEM, and SHV genes comprise the key forms of ESBL and are present in a wide range of clinically important pathogens globally [13].
Bangladesh, having suitable water bodies and comfortable weather, attracts a plethora of migratory birds mostly during the winter migration every year [14]. Migratory birds can contaminate these water bodies by transmitting pathogens via their fecal materials [15]. In Bangladesh, most of the habitats of migratory birds directly or indirectly interact with humans. Rural people are intimately associated with the water bodies which form the habitats of migratory birds and use them for activities such as fishing, bathing, and rearing ducks. In Bangladesh, most rural people rear ducks, chickens, and cattle and/or goats in their courtyards. In addition, they tend to use the contaminated water from these water bodies for agricultural purposes. Therefore, pathways exist for transmission of the pathogens of migratory birds to human and other animals. Thus, by contamination of water bodies, migratory birds pose a risk to both human and animal health.
Beta-lactamase-producing E. coli is often used to trace the evolution of multi-resistant bacteria in the environment and in migratory birds [5]. Several previous studies have indicated that migratory birds are carriers and reservoirs of ESBL-producing E. coli to humans and animals [4, 16]. In addition, they are suggested as carriers and reservoirs of antibiotic resistance genes globally [17, 18]. However, in Bangladesh, there is scarce data on the prevalence of ESBL-producing E. coli-carrying antibiotic resistance genes, and until now, the antibiotic resistance gene-carrying capabilities of migratory birds have been neglected. In the present study, we screened fecal materials of birds migrating via Bangladesh for ESBL-producing E. coli, to gain a better insight into the role of migratory birds as carriers of resistant bacteria, especially ESBL-producing E. coli.
Materials and Methods
Ethical Statement
The methodologies and related protocols used in this study were approved by the institutional ethical committee (AWEEC/BAU/2019(14)).
Selection of E. coli Isolates
E. coli strains used in this study were obtained in our previous study on migratory birds [19]. Original detection and isolation of these E. coli strains were done following culture on eosin methylene blue agar (HiMedia, India) plates, grams staining, biochemical tests (catalase test, coagulase test, sugar fermentation tests, methyl red test, Voges–Proskauer test, and indole test), and PCR targeting malB gene [19].
Detection of ESBL-Producing E. coli and Antimicrobial Susceptibility Testing
Antimicrobial resistance profiles of the isolated E. coli strains were determined by the Kirby–Bauer disk diffusion test [20]. Different classes of antibiotics, namely, penicillins (ampicillin, 25 µg), tetracyclines (tetracycline, 30 μg), fluoroquinolones (ciprofloxacin, 5 μg), aminoglycosides (gentamicin, 10 μg; streptomycin, 10 μg), macrolides (erythromycin, 15 μg), carbapenems (meropenem, 10 µg), polypeptides (colistin, 10 µg), and amphenicols (chloramphenicol, 10 μg) (HiMedia, India), were used in the sensitivity test. The results were interpreted in accordance with the guidelines of the Clinical and Laboratory Standards Institute [21]. ESBL-producing E. coli isolates were phenotypically screened by amoxicillin/clavulanate acid (amoxyclav) (30 µg), cefotaxime (30 µg), and ceftazidime (30 µg) disks (HiMedia, India) following the double-disk synergy test [21]. On Mueller–Hinton agar (HiMedia, India) plates inoculated with freshly growth bacterial culture (matched with 0.5 McFarland standard) (HiMedia, India), two third-generation cephalosporin-containing disks, namely, cefotaxime (30 μg) and ceftazidime (30 μg), were placed 30 mm apart (center to center) from an amoxicillin/clavulanic acid disk (amoxicillin, 20 μg, and clavulanic acid, 10 μg), followed by incubating in an incubator (SANYO, MCO175, Japan) for overnight at 37 °C and measuring the zone diameter of each disk. ESBL-producing E. coli were categorized as strains resistant to ceftazidime (≤ 17 mm) and cefotaxime (≤ 22 mm) and showed the enhancement of zone diameter (≥ 5 mm) of any of these two disks toward clavulanic acid containing disk [21]. Furthermore, any isolates showing resistance against three or more different classes of antibiotic were recognized as multidrug resistant (MDR) [22].
Detection of Beta-Lactamase Genes and Antibiotic Resistance Genes of E. coli Isolates
A simplex PCR assay was performed for the detection of beta-lactamase genes (blaTEM, blaCTX-M, blaCMY, and blaSHV) [23,24,25,26]. In addition, the genes associated with resistance to tetracyclines (tetA, tetB) [23], fluoroquinolones (qnrA, qnrB) [27, 28], aminoglycosides (aadA1) [23], macrolides (ereA) [24], and amphenicols (catA1) [24] were tested. Target genes and primers (Macrogen, Korea) associated with different beta-lactamase genes and antibiotic resistance genes are documented in Table 1.
For PCR, genomic DNA were extracted by boiling method as previously described [2]. Briefly, 1 ml of overnight freshly growth culture was centrifuged at 2,300 × g for 5 min in a centrifuge machine (KUBOTA 6500, Japan), and the supernatant was discarded. Then the pellet was suspended to 200 µl of phosphate buffer solution (PBS) (HiMedia, India) and boiled and cooled for 10 min in each step. After that, the suspension was centrifuged at 9,200 × g for 10 min, and the supernatant was collected as genomic DNA, followed by storing in Eppendorf tubes at − 20 °C for further use.
The PCR amplification was carried out with a 20 µl of final volume [master mix (2 ×) (Promega, Madison, WI, USA), 10 μL; nuclease-free water (Promega, Madison, WI, USA), 4 μL; each primer, 1 μL (0.2 pmol/ml); and genomic DNA (50 ng/ μL), 4 μL] in PCR thermal cycler (ASTEC, Japan). The thermal profile of PCR consisted initial denaturation, 95 °C for 5 min; 30 cycles of denaturation, 95 °C for 1 min; annealing variable temperature (Table 1) for 1 min; elongation, 72 °C for 1 min; and final extension, 72 °C for 10 min. PCR-positive controls consisted of E. coli genomic DNA which were previously positive for relevant genes. Non-template controls were used as PCR negative controls, where PBS was used instead of genomic DNA. The amplified PCR products were then analyzed with 1.5% agarose (Invitrogen, USA) in a gel electrophoresis apparatus (Nippon Genetics, Japan) and subsequently stained with ethidium bromide (0.5 μg/ml) (HiMedia, India). Finally, the ultra-violet trans-illuminator (Biometra, Germany) was used to capture the expected amplicon sizes. The sizes were compared with a 1 kb DNA ladder (Promega, Madison, WI, USA).
Statistical Analysis
Descriptive Analysis
Excel 2013 spreadsheet (Microsoft Office 2013, Microsoft, Los Angeles, CA, USA) was used to analyze data, and the Statistical Package for Social Science (SPSS) (IBM SPSS 25, IBM, Chicago, IL, USA) and GraphPad Prism version 8.4.3 (GraphPad Software, Inc.) were employed to perform statistical analysis.
The Pearson chi-square test for goodness-of-fit was performed to identify the variations among beta-lactamase genes of E. coli isolates. P-values less than 0.05 (p-value < 0.05) were deemed statistically significant. Following the Wilson/Brown hybrid method [29], binomial 95% confidence intervals were computed using GraphPad Prism.
Bivariate Analysis
Using IBM SPSS statistics (version 25), a bivariate analysis was employed to evaluate the correlation between antibiotics resistance to ESBL-producing E. coli and to determine the associations between beta-lactamase genes of E. coli isolates. Any p-value less than 0.05 was considered statistically significant.
Results
Phenotypic Prevalence of ESBL-Producing E. coli
Among 55 PCR-positive E. coli isolates, 21 (38.18%; 95% CI, 26.52–51.39%) were confirmed as phenotypically ESBL-producing E. coli by double-disk synergy test.
Genotypic Prevalence of Antibiotic-Resistant E. coli
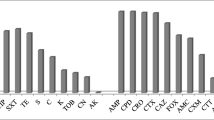
In the antibiogram, all the 55 E. coli isolates were found phenotypically resistant to ampicillin and erythromycin, 41 to streptomycin, 35 to tetracycline, 28 to ciprofloxacin, and 24 to chloramphenicol. By PCR, 90.24% (95% CI, 77.45–96.14%) of streptomycin-resistant isolates were found positive for the aadA1 gene; 100% (95% CI, 90.11–100.00%) and 31.43% (95% CI, 18.55–47.98%) of tetracycline-resistant isolates, respectively, positive for tetA and tetB genes; 35.71% (95% CI, 20.71–54.17%) and 25% (95% CI, 12.68–43.36%) of ciprofloxacin-resistant isolates positive for qnrA and qnrB genes, respectively; and all the erythromycin- and chloramphenicol-resistant isolates were negative for ereA and catA1 genes (Fig. 1).
Furthermore, of 21 ESBL-producing E. coli isolates, 20 isolates were positive for at least one antibiotic resistance gene. Among them, three isolates were positive for four resistance genes, followed by 11 for three genes and three for two or one gene. Notably, the tetA and aadA1 genes were present in 17 ESBL-producing E. coli isolates (Table 2).
CIP, ciprofloxacin; GEN, gentamicin; E, erythromycin; TE, tetracycline; MEM, meropenem; AMP, ampicillin; C, chloramphenicol; S, streptomycin.
Phenotypic MDR Nature of ESBL-Producing E. coli
All the ESBL-producing E. coli isolates displayed a MDR phenotype. In total, seven resistance patterns were identified among the ESBL-producing E. coli isolates. The most common MDR phenotype was ciprofloxacin, erythromycin, tetracycline, ampicillin, chloramphenicol, and streptomycin (CIP-E-TE-AMP-C-S) which was found in 11 (52.38%) of the ESBL-producing E. coli isolates. Furthermore, one isolate (BB-30) was resistant against 7 antibiotics (6 classes) (Table 2). By bivariate analysis, a highly significant correlation was observed between resistance patterns of ciprofloxacin and tetracycline (Pearson correlation coefficients, ρ = 0.868, p = < 0.001), ciprofloxacin and chloramphenicol (ρ = 0.791, p = < 0.001), and tetracycline and chloramphenicol (ρ = 0.686, p = 0.001). A moderately significant correlation was identified between tetracycline and meropenem (ρ = − 0.461, p = < 0.035) and streptomycin and meropenem (ρ = − 0.548, p = < 0.01) (Table 3).
A p-value less than 0.05 (p < 0.05) was deemed as significant. **Correlation is significant at the 0.01 level (2-tailed); *Correlation is significant at the 0.05 level (2-tailed); aCannot be computed because at least one of the variables is constant. CIP, ciprofloxacin; GEN, gentamicin; E, erythromycin; TE, tetracycline; MEM, meropenem; AMP, ampicillin; C, chloramphenicol; S, streptomycin.
Prevalence of Beta-Lactamase Genes in E. coli
Among the 21 ESBL-producing E. coli isolates, all harbored two or more beta-lactamase genes with blaTEM as the significantly dominant (chi-square test, p < 0.001) (20/21; 95.24%; 95% CI, 77.33–99.76%), followed by blaCMY (19/21; 90.48%; 95% CI, 71.09–98.31%), blaCTX-M (18/21; 85.71%; 95% CI, 65.36–95.02%), and blaSHV (9/21; 42.86%; 95% CI, 24.47–63.45%). The most common pattern was blaTEM–blaCMY–blaCTX-M presenting in nine ESBL-producing E. coli isolates. In addition, seven isolates were found positive for all four beta-lactamase genes (blaTEM–blaCMY–blaCTX-M–blaSHV) and four for any of two genes (Table 2).
Comparison of Pearson Correlation Coefficients Among ESBL-Producing E. coli
The Pearson correlation coefficients (ρ) derived from bivariate statistical analysis revealed a highly significant correlation between the blaTEM and blaCMY beta-lactamase genes of E. coli (ρ = 0.689; p = 0.001). However, the other genes did not show any significant correlation (p > 0.05) (Table 4).
Here, a p-value less than 0.05 was deemed statistically significant. **Correlation is significant at the 0.01 level (two-tailed).
Discussion
There is an acknowledged failure to detect antibiotic resistance and the associated genes from migratory birds in Bangladesh. Although several studies were conducted previously in Bangladesh to detect ESBL-producing E. coli [30,31,32], until now their association with migratory birds has not been routinely reported. In this study, we therefore reported antibiotic resistance and beta-lactamase genes in E. coli isolated from migratory birds travelling to Bangladesh.
The E. coli strains isolated from migratory birds harbored several resistance genes including tetA, tetB, qnrA, qnrB, and aadA1 genes; of these, tetA was found in all tetracycline-resistant phenotypes. However, there was an absence of ereA and catA1 genes in the E. coli isolates. Previously, Dolejska et al. [33] detected the high occurrence of different resistance genes from black-headed gulls in the Czech Republic. In addition, Radhouani et al. [34] recorded several antibiotic resistance genes from migratory birds, which is consistent with the present study. The occurrence of resistant E. coli in migratory birds may be associated with the transmission of antibiotic resistance genes to other environments. Water contaminated with feces of migratory birds could be a significant risk factor for the dissemination of resistant E. coli pathogens and their resistance genes in the environments. In addition, the spread of E. coli-carrying antibiotic resistance genes from migratory birds to humans and animals can occur via contaminated fresh and seawater systems [35].
The near pandemic spreading of ESBL-producing bacteria is of great public health concern across the globe. The occurrence of ESBL-producing E. coli has been increasing in many countries, not only restricted to humans but also reported in environmental niches like migratory birds, livestock, water, and in soils [32]. The present study indicates that many migratory birds (38.18%) are carriers of ESBL-producing E. coli in Bangladesh. This high prevalence is comparable to the previous studies detecting ESBL-producing E. coli in 30% of wild ducks [30] and in 17.3% of migratory gulls [31] in Bangladesh and in 17% of wild migratory birds in Pakistan [18]. These results and the data presented here indicate that migratory birds play a significant role in transmitting and spreading ESBL-producing E. coli in Asia via the Indus migration route. Globally, multiple studies detected ESBL-producing E. coli [4, 8, 36,37,38], illustrating the global importance of migratory birds as potential carriers and reservoirs. The results found in the present study suggest that migratory birds may contribute to the dissemination of ESBL-producing E. coli in Bangladesh.
In the current study, all the ESBL-producing E. coli isolates were phenotypically MDR in nature. Previously, Mohsin et al. [18] detected MDR ESBL-producing E. coli from 88.84% of wild migratory birds travelling to Pakistan. Here, highly significant positive correlations between resistance patterns of tetracycline and ciprofloxacin, chloramphenicol and ciprofloxacin, and chloramphenicol and streptomycin were shown by bivariate analysis. These significant associations might be due to the hazardous use of antimicrobial agents in areas usually inhabited by migratory birds. Environmental contamination might also play a pivotal role in such cases. The high occurrence of MDR ESBL-producing E. coli from our study is of great concern and may be due to the deterioration of the surrounding environments.
In the present study, blaTEM was detected at a predominant rate compared to other ESBL genotypes, as previously reported [37]. In addition, ESBL genotypes blaCMY and blaCTX-M were also frequently detected in our study. The beta-lactamase gene blaSHV was detected in more than 40% of ESBL-producing E. coli isolates. These results are in agreement with previous studies [8, 18, 30,31,32, 38, 39]. ESBL genotypes blaCTX-M are increasingly distributed in humans, animals, and environmental sources which is revealing an urgent problem in infectious disease treatment [40]. The genes blaCTX-M are very common in wild birds and are generally found in human- and veterinary-origin ESBL isolates [41]. Though the AmpC-type beta-lactamase gene blaCMY-producing E. coli is commonly associated with humans, companion, and food-producing animals, it is also commonly detected in avian wildlife [40]. In addition to blaCMY and blaCTX-M, the beta-lactamase genes blaTEM and blaSHV also confer resistance to beta-lactam classes of antibiotics in humans, livestock, and other animals [37]. The high prevalence of beta-lactamase genes in E. coli isolated from migratory birds poses a serious threat to human health. This is because migratory birds are directly connected to environmental features, especially water, and contaminated water plays a significant role in the dissemination of beta-lactamase-producing E. coli in the human community.
In this study, we found that 95.24% ESBL-producing E. coli harbored one or more antibiotic resistance genes along with beta-lactamase genes. Previously, Mohsin et al. [18] detected similar resistance genes in ESBL-producing E. coli isolates from migratory birds in Pakistan. The occurrence of MDR ESBL-producing E. coli isolates in the migratory birds travelling to Bangladesh in winter is alarming, as possible consequences would be severe clinical outcomes concomitant with serious limitations in antimicrobial treatment.
To the best of our knowledge, this is the first study in Bangladesh to detect E. coli-carrying beta-lactamase blaCMY and blaSHV genes from migratory birds; however, the study has several limitations. Here, we did not focus on environmental samples contaminated with the migratory birds’ fecal materials to confirm transmission of AMR from these birds to the environment. In addition, a limited number of samples were analyzed in this study. A further detailed study comprising more samples and in-depth genetic analysis of the resistant isolates could have been more informative.
Conclusions
The present study confirms the detection of ESBL-producing E. coli harboring clinically important blaCMY and blaSHV and other resistance genes from migratory birds for the first time in Bangladesh. The findings indicate that migratory birds are potential carriers and spreaders of AMR- and ESBL-producing organisms in humans, animals, and vulnerable environments in Bangladesh and pose a serious threat to one-health components. Therefore, these birds need to be kept under antibiotic resistance surveillance for better management of AMR-related hazards in Bangladesh.
References
Tawyabur M, Islam M, Sobur M, Hossain M, Mahmud M, Paul S, Hossain MT, Ashour HM, Rahman M (2020) Isolation and characterization of multidrug-resistant Escherichia coli and Salmonella spp. from healthy and diseased turkeys. Antibiotics 9:770. https://doi.org/10.3390/antibiotics9110770.
Ievy S, Islam M, Sobur M, Talukder M, Rahman M, Khan MF, Rahman M (2020) Molecular detection of avian pathogenic Escherichia coli (APEC) for the first time in layer farms in Bangladesh and their antibiotic resistance patterns. Microorganisms 8:1021. https://doi.org/10.3390/microorganisms8071021
Oteo J, Mencia A, Bautista V, Pastor N, Lara N, Gonzalez-Gonzalez F, García-Peña FJ (2018) Campos J (2018) Colonization with Enterobacteriaceae-producing ESBLs, AmpCs, and OXA-48 in wild avian species, Spain 2015–2016. Microb Drug Resist 24:932–938. https://doi.org/10.1089/mdr.2018.0004
Zurfluh K, Albini S, Mattmann P, Kindle P, Nüesch-Inderbinen M, Stephan R, Vogler BR (2019) Antimicrobial resistant and extended-spectrum β-lactamase producing Escherichia coli in common wild bird species in Switzerland. MicrobiologyOpen 8:e845. https://doi.org/10.1002/mbo3.845
Guenther S, Ewers C, Wieler LH (2011) Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol 19(2):246. https://doi.org/10.3389/fmicb.2011.00246
Rahman M, Sobur M, Islam M, Ievy S, Hossain M, El Zowalaty ME, Rahman AM, Ashour HM (2020) Zoonotic diseases: etiology, impact, and control. Microorganisms 8:1405. https://doi.org/10.3390/microorganisms8091405
Haberecht HB, Nealon NJ, Gilliland JR, Holder AV, Runyan C, Oppel RC, Ibrahim HM, Mueller L, Schrupp F, Vilchez S, Antony L (2019) Antimicrobial-resistant Escherichia coli from environmental waters in northern Colorado. J Environ Public Health 2019:3862949. https://doi.org/10.1155/2019/3862949
Parker D, Sniatynski MK, Mandrusiak D, Rubin JE (2016) Extended-spectrum β-lactamase producing Escherichia coli isolated from wild birds in Saskatoon, Canada. Lett Appl Microbiol 63:11–15. https://doi.org/10.1111/lam.12589
Bryskier A (2005) Penicillins. In: Bryskier A (ed) Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC, pp 113–162
Pitout JD (2010) Infections with extended-spectrum β-lactamase-producing Enterobacteriaceae. Drugs 70:313–333. https://doi.org/10.2165/11533040-000000000-00000
Pehlivanlar Önen S, Aslantaş Ö, Şebnem Yılmaz E, Kürekci C (2015) Prevalence of β-lactamase producing Escherichia coli from retail meat in Turkey. J Food Sci 80:M2023–M2029. https://doi.org/10.1111/1750-3841.12984
Pitout JD, Laupland KB (2008) Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. https://doi.org/10.1016/S1473-3099(08)70041-0
Paterson DL, Bonomo RA (2005) Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. https://doi.org/10.1128/CMR.18.4.657-686.2005
Akter M, Islam MS, Islam MA, Sobur MA, Jahan MS, Rahman S, Nazir KN, Rahman MT (2020) Migratory birds as the potential source for the transmission of Aspergillus and other fungus to Bangladesh. J Adv Vet Anim Res 7:338–344. https://doi.org/10.5455/javar.2020.g427
Zhao G, Zhou L, Dong Y, Cheng Y, Song Y (2017) The gut microbiome of hooded cranes (Grus monacha) wintering at Shengjin Lake. China Microbiologyopen 6:e00447. https://doi.org/10.1002/mbo3.447
Blaak H, Lynch G, Italiaander R, Hamidjaja RA, Schets FM, de Roda Husman AM (2015) Multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLoS ONE 10:e0127752. https://doi.org/10.1371/journal.pone.0127752
Alcalá L, Alonso CA, Simón C, González-Esteban C, Orós J, Rezusta A, Ortega C, Torres C (2016) Wild birds, frequent carriers of extended-spectrum β-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb Ecol 72:861–869. https://doi.org/10.1007/s00248-015-0718-0
Mohsin M, Raza S, Schaufler K, Roschanski N, Sarwar F, Semmler T, Schierack P, Guenther S (2017) High prevalence of CTX-M-15-Type ESBL-producing E. coli from migratory avian species in Pakistan. Front Microbial 12(8):2476. https://doi.org/10.3389/fmicb.2017.02476
Islam M, Nayeem M, Hasan M, Sobur M, Ievy S, Rahman S, Kafi M, Ashour HM, Rahman M (2021) Virulence determinants and multidrug resistance of Escherichia coli isolated from migratory birds. Antibiotics 10:190. https://doi.org/10.3390/antibiotics10020190
Bayer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol 45:493–496
CLSI (2018) Performance standards for antimicrobial susceptibility testing, 28th ed.; CLSI Supplement M100s; Clinical and Laboratory Standards Institute: Wayne, PA, USA.
Sweeney MT, Lubbers BV, Schwarz S, Watts JL (2018) Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother 73:1460–1463. https://doi.org/10.1093/jac/dky043
Randall LP, Cooles SW, Osborn MK, Piddock LJ, Woodward MJ (2004) Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother 53:208–216. https://doi.org/10.1093/jac/dkh070
Van TT, Chin J, Chapman T, Tran LT, Coloe PJ (2008) Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol 124:217–223. https://doi.org/10.1016/j.ijfoodmicro.2008.03.029
Gundran RS, Cardenio PA, Villanueva MA, Sison FB, Benigno CC, Kreausukon K, Pichpol D, Punyapornwithaya V (2019) Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended-spectrum β-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet Res 15:1–8. https://doi.org/10.1186/s12917-019-1975-9
Mandakini R, Roychoudhury P, Subudhi PK, Kylla H, Samanta I, Bandyopadhayay S, Dutta TK (2020) Higher prevalence of multidrug-resistant extended-spectrum β-lactamases producing Escherichia coli in unorganized pig farms compared to organized pig farms in Mizoram, India. Vet World 13(12):2752–2758. https://doi.org/10.14202/vetworld.2020.2752-2758
Shahrani M, Dehkordi FS, Momtaz H (2014) Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol Res 47:1–13. https://doi.org/10.1186/0717-6287-47-28
Wang A, Yang Y, Lu Q, Wang Y, Chen Y, Deng L, Ding H, Deng Q, Zhang H, Wang C, Liu L (2008) Presence of qnr gene in Escherichia coli and Klebsiella pneumoniae resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infect Dis 8:1–6. https://doi.org/10.1186/1471-2334-8-68
Brown LD, Cai TT, DasGupta A (2001) Interval estimation for a binomial proportion. Stat Sci 16:101–117
Hasan B, Sandegren L, Melhus Å, Drobni M, Hernandez J, Waldenström J, Alam M, Olsen B (2012) Antimicrobial drug–resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg Infect Dis 18:2055–2058. https://doi.org/10.3201/eid1812.120513
Hasan B, Melhus Å, Sandegren L, Alam M, Olsen B (2014) The gull (Chroicocephalus brunnicephalus) as an environmental bioindicator and reservoir for antibiotic resistance on the coastlines of the Bay of Bengal. Microb Drug Resist 20:466–471. https://doi.org/10.1089/mdr.2013.0233
Rashid M, Rakib MM, Hasan B (2015) Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect Ecol Epidemiology 5:26712. https://doi.org/10.3402/iee.v5.26712
Dolejska M, Cizek A, Literak I (2007) High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J Appl Microbiol 103:11–19. https://doi.org/10.1111/j.1365-2672.2006.03241.x
Radhouani H, Poeta P, Goncalves A, Pacheco R, Sargo R, Igrejas G (2012) Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). J Med Microbiol 61:837–843. https://doi.org/10.1099/jmm.0.038364-0
Ahmed ZS, Elshafiee EA, Khalefa HS, Kadry M, Hamza DA (2019) Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob Resist Infect Control 8:1–8. https://doi.org/10.1186/s13756-019-0657-5
Sandegren L, Stedt J, Lustig U, Bonnedahl J, Andersson DI, Järhult JD (2018) Long-term carriage and rapid transmission of extended spectrum beta-lactamase-producing E. coli within a flock of Mallards in the absence of antibiotic selection. Environ Microbiol Rep 10:576–582. https://doi.org/10.1111/1758-2229.12681
Ngaiganam EP, Pagnier I, Chaalal W, Leangapichart T, Chabou S, Rolain JM, Diene SM (2019) Investigation of urban birds as source of β-lactamase-producing Gram-negative bacteria in Marseille city, France. Acta Vet Scand 61:1–7. https://doi.org/10.1186/s13028-019-0486-9
Darwich L, Vidal A, Seminati C, Albamonte A, Casado A, López F, Molina-López RA, Migura-Garcia L (2019) High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 14:e0210686. https://doi.org/10.1371/journal.pone.0210686
Veldman K, van Tulden P, Kant A, Testerink J, Mevius D (2013) Characteristics of cefotaxime-resistant Escherichia coli from wild birds in the Netherlands. Appl Environ Microbiol 79:7556–7561. https://doi.org/10.1128/AEM.01880-13
Wang J, Ma ZB, Zeng ZL, Yang XW, Huang Y, Liu JH (2017) The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool Res 38(2):55–80. https://doi.org/10.24272/j.issn.2095-8137.2017.003
Pietsch M, Eller C, Wendt C, Holfelder M, Falgenhauer L, Fruth A, Grössl T, Leistner R, Valenza G, Werner G, Pfeifer Y (2017) Molecular characterisation of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet Microbial 200:130–137. https://doi.org/10.1016/j.vetmic.2015.11.028
Acknowledgements
We would like to thank Dr. Jayedul Hassan, Department of Microbiology and Hygiene, Bangladesh Agricultural University, for providing the PCR-positive controls. We also thank to Dr. AMM Taufiquer Rahman, Zilla Hospital, Naogaon, Bangladesh, for comments and suggestions on this this study.
Funding
We would like to thank the Bangladesh Agricultural University Research System (BAURES; 2019/8/BAU) and University Grants Commission of Bangladesh (2020/28/UGC) for facilitating parts of the study.
Author information
Authors and Affiliations
Contributions
Conceptualization, MSI, and MTR; methodology, MSI; software, MSI; validation, MTR; formal analysis, MSI, MAS, SI, and MTR; investigation, MSI, MPS and SI; data curation, MSI; writing—original draft preparation, MSI, MAS, and MTR; writing—review and editing, MSI, MAS, MR, FMB, MPS, and MTR; supervision, MAK and MTR; project administration, SR and MTR; critical revisions and writing, MTR.
Corresponding author
Ethics declarations
Ethics Approval
The methodologies and related protocols used in this study were approved by the institutional ethical committee (AWEEC/BAU/2019(14)).
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Islam, M.S., Sobur, M.A., Rahman, S. et al. Detection of blaTEM, blaCTX-M, blaCMY, and blaSHV Genes Among Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Isolated from Migratory Birds Travelling to Bangladesh. Microb Ecol 83, 942–950 (2022). https://doi.org/10.1007/s00248-021-01803-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01803-x