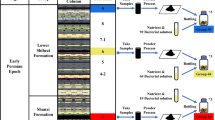
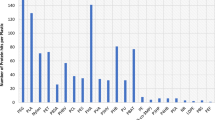
Abstract
This article presents the first experimental data on the ability of microbial communities from sediments of the Gorevoy Utes natural oil seep to degrade petroleum hydrocarbons under anaerobic conditions. Like in marine ecosystems associated with oil discharge, available electron acceptors, in particular sulfate ions, affect the composition of the microbial community and the degree of hydrocarbon conversion. The cultivation of the surface sediments under sulfate-reducing conditions led to the formation of a more diverse bacterial community and greater loss of n-alkanes (28%) in comparison to methanogenic conditions (6%). Microbial communities of both surface and deep sediments are more oriented to degrade polycyclic aromatic hydrocarbons (PAHs), to which the degree of the PAH conversion testifies (up to 46%) irrespective of the present electron acceptors. Microorganisms with the uncultured closest homologues from thermal habitats, sediments of mud volcanoes, and environments contaminated with hydrocarbons mainly represented microbial communities of enrichment cultures. The members of the phyla Firmicutes, Chloroflexi, and Caldiserica (OP5), as well as the class Deltaproteobacteria and Methanomicrobia, were mostly found in enrichment cultures. The influence of gas-saturated fluids may be responsible for the presence in the bacterial 16S rRNA gene libraries of the sequences of “rare taxa”: Planctomycetes, Ca. Atribacteria (OP9), Ca. Armatimonadetes (OP10), Ca. Latescibacteria (WS3), Ca. division (AC1), Ca. division (OP11), and Ca. Parcubacteria (OD1), which can be involved in hydrocarbon oxidation.








Similar content being viewed by others
References
Augustinovic Z, Birketveit O, Clements K, Freeman M, Gopi S, Ishoey T, Jackson G, Kubala G, Larsen J, Marcotte B, Scheie J, Skovhus T, Sunde E (2012) Microbes –oilfield enemies or allies? Oilfield Rev 24:4–17
Roling WFM, Head IM, Larter SR (2003) The microbiology of hydrocarbon degradation in subsurface petroleum reservoirs: perspectives and prospects. Res Microbiol 154:321–328. https://doi.org/10.1016/S0923-2508(03)00086-X
Head IM, Larter SR, Gray ND, Sherry A, Adams JJ, Aitken CM, Jones DM, Rowan AK, Huang H, Roling WFM (2010) Hydrocarbon degradation in petroleum reservoirs. In: Timmis KN (ed) Handbook of Hydrocarbon and Lipid Microbiology. Springer, Berlin, pp 3098–3110. https://doi.org/10.1007/978-3-540-77587-4_232
Gieg LM, Davidova IA, Duncan KE, Suflita JM (2010) Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ Microbiol 12:3074–3086. https://doi.org/10.1111/j.1462-2920.2010.02282.x
Bian XY, Mbadinga SM, Liu YF, Yang SZ, Liu JF, Ye RQ, Gu JD, Mu BZ (2015) Insights into the anaerobic biodegradation pathway of n-alkanes in oil reservoirs by detection of signature metabolites. Sci Rep 5:9801. https://doi.org/10.1038/srep09801
Stagars MH, Ruff SE, Amann R, Knittel K (2016) High diversity of anaerobic alkane-degrading microbial communities in marine seep sediments based on (1-methylalkyl)succinate synthase genes. Front Microbiol 6:1511. https://doi.org/10.3389/fmicb.2015.01511
Cheng L, Shi SB, Yang L, Zhang YH, Dolfing J, Sun YG, Liu LY, Li Q, Tu B, Dai LR, Shi Q, Zhang H (2019) Preferential degradation of long-chain alkyl substituted hydrocarbons in heavy oil under methanogenic conditions. Org Geochem 138:103927. https://doi.org/10.1016/j.orggeochem.2019.103927
Pannekens M, Kroll L, Muller H, Mbow FT, Meckenstock RU (2019) Oil reservoirs, an exceptional habitat for microorganisms. New Biotechnol 49:1–9. https://doi.org/10.1016/j.nbt.2018.11.006
Ji JH, Liu YF, Zhou L, Mbadinga SM, Pan P, Chen J, Liu JF, Yang SZ, Sand W, Gu JD, Mu BZ (2019) Methanogenic degradation of long n-alkanes requires fumarate-dependent activation. Appl Environ Microbiol 85:e00985–e00919. https://doi.org/10.1128/aem.00985-19
Laczi K, Kis AE, Szilagyi A, Bounedjoum N, Bodor A, Vincze GE, Kovacs T, Rakhely G, Perei K (2020) New frontiers of anaerobic hydrocarbon biodegradation in the multi-omics era. Front Microbiol 11:590049. https://doi.org/10.3389/fmicb.2020.590049
Sierra-Garcia IN, Dellagnezze BM, Santos VP, Chaves MR, Capilla R, Neto EVS, Gray N, Oliveira VM (2017) Microbial diversity in degraded and non-degraded petroleum samples and comparison across oil reservoirs at local and global scales. Extremophiles 21:211–229. https://doi.org/10.1007/s00792-016-0897-8
Dong XY, Greening C, Rattray JE, Chakraborty A, Chuvochina M, Mayumi D, Dolfing J, Li C, Brooks JM, Bernard BB, Groves RA, Lewis IA, Hubert CRJ (2019) Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat Commun 10:1816. https://doi.org/10.1101/400804
McIntosh P, Schulthess CP, Kuzovkina YA, Guillard K (2017) Bioremediation and phytoremediation of total petroleum hydrocarbons (TPH) under various conditions. Int J Phytoremediation 19:755–764. https://doi.org/10.1080/15226514.2017.1284753
Zhang K, Hu Z, Zeng FF, Yang XY, Wang JJ, Jing R, Zhang HN, Li YT, Zhang Z (2019) Biodegradation of petroleum hydrocarbons and changes in microbial community structure in sediment under nitrate-, ferric-, sulfate-reducing and methanogenic conditions. J Environ Manag 249:109425. https://doi.org/10.1016/j.jenvman.2019.109425
Simoneit BRT, Aboul-Kassim TAT, Tiercelin JJ (2000) Hydrothermal petroleum from lacustrine sedimentary organic matter in the East African Rift. Appl Geochem 15:355–368. https://doi.org/10.1016/s0883-2927(99)00044-x
Zarate-del Valle PF, Rushdi AI, Simoneit BRT (2006) Hydrothermal petroleum of Lake Chapala, Citala Rift, western Mexico: Bitumen compositions from source sediments and application of hydrous pyrolysis. Appl Geochem 21:701–712. https://doi.org/10.1016/j.apgeochem.2006.01.002
Kontorovich AE, Kashirtsev VA, Moskvin VI, Burshtein LM, Zemskaya TI, Kostyreva EA, Kalmychkov GV, Khlystov OM (2007) Petroleum potential of Baikal deposits. Russ Geol Geophys 48:1046–1053. https://doi.org/10.1016/j.rgg.2007.11.004
Khlystov OM, Gorshkov AG, Egorov AV, Zemskaya TI, Granin NG, Kalmychkov GV, Vorob ' eva SS, Pavlova ON, Yakup MA, Makarov MM, Moskvin VI, Grachev MA (2007) Oil in the lake of world heritage. Dokl Earth Sci (Moscow) 415:682–685. https://doi.org/10.1134/S1028334X07050042
Gorshkov A, Pavlova O, Khlystov O, Zemskaya T (2020) Fractioning of petroleum hydrocarbons from seeped oil as a factor of purity preservation of water in Lake Baikal (Russia). J Great Lakes Res 46:115–122. https://doi.org/10.1016/j.jglr.2019.10.010
Khlystov OM, Zemskaya TI, Sitnikova TY, Mekhanikova IV, Kaigorodova IA, Gorshkov AG, Timoshkin OA, Shubenkova OV, Chernitsyna SM, Lomakina AV, Likhoshvai AV, Sagalevich AM, Moskvin VI, Peresypkin VI, Belyaev NA, Slipenchuk MV, Tulokhonov AK, Grachev MA (2009) Bottom bituminous constructions and biota inhabiting them according to investigation of Lake Baikal with the Mir submersible. Dokl Earth Sci 429:1333–1336
Sitnikova TY, Zemskaya TI, Chernitsina SM, Likhoshway AV, Klimenkov IV, Naumova TV (2015) Structure of biocenoses formed on bitumen mounds in the abyssal zone of Lake Baikal. Russ J Ecol 3:292–298. https://doi.org/10.1134/S106741361503011X
Pavlova ON, Lomakina AV, Gorshkov AG, Suslova MY, Likhoshvai AV, Zemskaya TI (2012) Microbial communities and their ability to oxidize n-alkanes in the area of release of gas- and oil-containing fluids in Mid-Baikal (Cape Gorevoi Utes). Biol Bull (Moscow) 39:458–463. https://doi.org/10.1134/s1062359012050123
Pavlova ON, Izosimova ON, Chernitsyna SM, Ivanov VG, Pogodaeva TV, Gorchkov AG (2020) Processes of anaerobic oxidation of oil in bottom sediments of Lake Baikal. Limnol Freshw Biol 4:1006–1007. https://doi.org/10.31951/2658-3518-2020-A-4-1006
Widdel F, Pfennig N (1982) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch Microbiol 131:360–365. https://doi.org/10.1007/bf00406470
Netrusov A (2005) Microbiological practicum. Student handbook. Academia Publ House, Moscow, p 612 (in Russian)
Mizandrontsev IB, Kozlov VV, Ivanov VG, Kucher KM, Korneva ES, Granin NG (2020) Vertical distribution of methane in Baikal water. Water Res (Moscow) 47:122–129. https://doi.org/10.1134/s0097807820010108
Pogodaeva TV, Zemskaya TI, Golobokova LP, Khlystov OM, Minami H, Sakagami H (2007) Chemical composition of pore waters of bottom sediments in different Baikal basins. Russ Geol Geophys 48:886–900. https://doi.org/10.1016/j.rgg.2007.02.012
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A Laboratory Manual. Cold Spring Harbor, New York, p 545
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, pp 115–175
Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF (2003) Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl Environ Microbiol 69:5483–5491. https://doi.org/10.1128/aem.69.9.5483-5491.2003
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/aem.01541-09
Gorshkov AG, Izosimova ON, Pavlova ON, Khlystov ON, Zemskaya TI (2020) Assessment of water pollution near the deep oil seep in Lake Baikal. Limnol Freshw Biol 2:397–404. https://doi.org/10.31951/2658-3518-2020-A-2-397
Pogodaeva TV, Lopatina IN, Khlystov OM, Egorov AV, Zemskaya TI (2017) Background composition of pore waters in Lake Baikal bottom sediments. J Great Lakes Res 43:1030–1043. https://doi.org/10.1016/j.jglr.2017.09.003
Rissanen AJ, Peura S, Mpamah PA, Taipale S, Tiirola M, Biasi C, Mäki A, Nykänen H (2019) Vertical stratification of bacteria and archaea in sediments of a small boreal humic lake. FEMS Microbiol Lett 366:fnz044. https://doi.org/10.1093/femsle/fnz044
Zhou Z, Pan J, Wang F, Gu J-D, Li M (2018) Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol Rev 42:639–655. https://doi.org/10.1093/femsre/fuy023
Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG (2017) Archaea and the origin of eukaryotes. Nat Rev Microbiol 15:711–723. https://doi.org/10.1038/nrmicro.2017.133
Mori K, Sunamura M, Yanagawa K, Ishibashi J-I, Miyoshi Y, Iino T, Suzuki K-I, Urabe T (2008) First cultivation and ecological investigation of a bacterium affiliated with the Candidate Phylum OP5 from hot springs. Appl Environ Microbiol 74:6223–6229. https://doi.org/10.1128/aem.01351-08
Youssef NH, Farag IF, Rinke C, Hallam SJ, Woyke T, Elshahed MS (2015) In silico analysis of the metabolic potential and niche specialization of candidate phylum “Latescibacteria” (WS3). PLoS One 10:e0127499. https://doi.org/10.1371/journal.pone.0127499
Lee YJ, Romanek CS, Mills GL, Davis RC, Whitman WB, Wiegel J (2006) Gracilibacter thermotolerans gen. nov., sp. nov., an anaerobic, thermotolerant bacterium from a constructed wetland receiving acid sulfate water. Int J Syst Evol Microbiol 56:2089–2093. https://doi.org/10.1099/ijs.0.64040-0
Kragelund C, Levantesi C, Borger A, Thelen K, Eikelboom D, Tandoi V, Kong Y, Van Der Waarde J, Krooneman J, Rossetti S, Thomsen TR, Nielsen PH (2007) Identity, abundance and ecophysiology of filamentous Chloroflexi species present in activated sludge treatment plants. FEMS Microbiol Ecol 59:671–682. https://doi.org/10.1111/j.1574-6941.2006.00251.x
Kadnikov VV, Mardanov AV, Beletsky AV, Karnachuk OV, Ravin NV (2019) Genome of the candidate phylum Aminicenantes bacterium from a deep subsurface thermal aquifer revealed its fermentative saccharolytic lifestyle. Extremophiles 23:189–200. https://doi.org/10.1007/s00792-018-01073-5
Hu P, Tom L, Singh A, Thomas BC, Baker BJ, Piceno YM, Andersen GL, Banfield JF (2016) Genome-resolved metagenomic analysis reveals roles for candidate phyla and other microbial community members in biogeochemical transformations in oil reservoirs. MBio 7:e01669–e01615. https://doi.org/10.1128/mbio.01669-15
Dombrowski N, Seitz KW, Teske AP, Baker BJ (2017) Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 5:106. https://doi.org/10.1186/s40168-017-0322-2
Zhang C, Meckenstock RU, Weng S, Wei G, Hubert CRJ, Wang J-H, Dong X (2020) Marine sediments harbor diverse archaea and bacteria with potentials for anaerobic hydrocarbon degradation via fumarate addition. Posted Content published. https://doi.org/10.21203/rs.3.rs-71489/v1
Zemskaya TI, Bukin SV, Lomakina AV, Pavlova ON (2021) Microorganisms in bottom sediments of Lake Baikal, the world’s deepest and oldest lake. Microbiology (Moscow) 90:298–313. https://doi.org/10.1134/S0026261721030140
Namsaraev BB, Zemskaya TI (2000) Microbiological processes of the carbon cycle in bottom sediments of Lake Baikal. Geo, Novosibirsk, p 160 (In Russian)
Pavlova ON, Bukin SV, Lomakina AV, Kalmychkov GV, Ivanov VG, Morozov IV, Pogodaeva TV, Pimenov NV, Zemskaya TI (2014) Production of gaseous hydrocarbons by microbial communities of Lake Baikal bottom sediments. Microbiology (Moscow) 83:798–804. https://doi.org/10.1134/s0026261714060137
Bukin SV, Pavlova ON, Kalmychkov GV, Ivanov VG, Pogodaeva TV, Galachyants YP, Bukin YS, Khabuev AV, Zemskaya TI (2018) Substrate specificity of methanogenic communities from Lake Baikal bottom sediments associated with hydrocarbon gas discharge. Microbiology (Moscow) 87:549–558. https://doi.org/10.1134/s0026261718040045
Rabus R, Boll M, Heider J, Meckenstock RU, Buckel W, Einsle O, Ermler U, Golding BT, Gunsalus RP, Kroneck PMH, Krüger M, Lueders T, Martins BM, Musat F, Richnow HH, Schink B, Seifert J, Szaleniec M, Treude T, Ullmann GM, Vogt C, von Bergen M, Wilkes H (2016) Anaerobic microbial degradation of hydrocarbons: from enzymatic reactions to the environment. J Mol Microbiol Biotechnol 26:5–28. https://doi.org/10.1159/000443997
Tan BF, Semple K, Foght J (2015) Anaerobic alkane biodegradation by cultures enriched from oil sands tailings ponds involves multiple species capable of fumarate addition. FEMS Microbiol Ecol 91:fiv042. https://doi.org/10.1093/femsec/fiv042
Boopathy R (2004) Anaerobic biodegradation of no. 2 diesel fuel in soil: a soil column study. Bioresour Technol 94:143–151. https://doi.org/10.1016/j.biortech.2003.12.006
Falkner KK, Measures CI, Herbelin SE, Edmond JM, Weiss RF (1991) The major and minor element geochemistry of Lake Baikal. Limnol Oceanogr 36:413–423. https://doi.org/10.4319/lo.1991.36.3.0413
Ma TT, Liu LY, Rui JP, Yuan Q, Feng DS, Zhou Z, Dai LR, Zeng WQ, Zhang H, Cheng L (2017) Coexistence and competition of sulfate-reducing and methanogenic populations in an anaerobic hexadecane-degrading culture. Biotechnol Biofuels 10:207. https://doi.org/10.1186/s13068-017-0895-9
Pavlova ON, Izosimova ON, Gorshkov AG, Novikova AS, Bukin SV, Ivanov VG, Khlystov OM, Zemskaya TI (2020) Current state of deep oil seepage near cape Gorevoi Utes (Central Baikal). Russ Geol Geophys 61:1007–1014. https://doi.org/10.15372/rgg2019180
Pimenov NV, Zakharova EE, Bryukhanov AL, Korneeva VA, Kuznetsov BB, Tourova TP, Pogodaeva TV, Kalmychkov GV, Zemskaya TI (2014) Activity and structure of the sulfate-reducing bacterial community in the sediments of the southern part of Lake Baikal. Microbiology (Moscow) 83:47–55. https://doi.org/10.1134/s0026261714020167
Gray ND, Sherry A, Hubert C, Dolfing J, Head IM (2010) Methanogenic degradation of petroleum hydrocarbons in subsurface environments: remediation, heavy oil formation, and energy recovery. In: Laskin AI, Sariaslani S, Gadd GM (eds) Adv Appl Microbiol. Academic Press, pp 137–161. https://doi.org/10.1016/s0065-2164(10)72005-0
Wentzel A, Lewin A, Cervantes FJ, Valla S, Kotlar HK (2013) Deep subsurface oil reservoirs as poly-extreme habitats for microbial life. A current review. In: Seckbach J, Oren A, Stan-Lotter H (eds) Polyextremophiles. Cellular Origin, Life in Extreme Habitats and Astrobiology. Springer, Dordrecht, pp 439–466. https://doi.org/10.1007/978-94-007-6488-0_19
Kleinsteuber S, Schleinitzm KM, Vogt C (2012) Key players and team play: anaerobic microbial communities in hydrocarbon-contaminated aquifers. Appl Microbiol Biotechnol 94:851–873. https://doi.org/10.1007/s00253-012-4025-0
Zemskaya TI, Lomakina AV, Mamaeva EV, Zakharenko AS, Pogodaeva TV, Petrova DP, Galachyants YP (2015) Bacterial communities in sediments of Lake Baikal from areas with oil and gas discharge. Aquat Microb Ecol 76:95–109. https://doi.org/10.3354/ame01773
Lomakina AV, Pogodaeva TV, Morozov IV, Zemskaya TI (2014) Microbial communities of the discharge zone of oil and gas-bearing fluids in low-mineral Lake Baikal. Microbiology 83:278–287. https://doi.org/10.1134/s0026261714030126
Kadnikov VV, Mardanov A, Beletsky AV, Shubenkova OV, Pogodaeva TN, Zemskaya TI, Ravin NV, Skryabin KG (2012) Microbial community structure in methane hydrate-bearing sediments of freshwater Lake Baikal. FEMS Microbiol Ecol 79:348–358. https://doi.org/10.1111/j.1574-6941.2011.01221.x
Chakraborty A, Ruff SE, Dong X, Ellefson ED, Li C, Brooks JM, McBee J, Bernard BB, Hubert CRJ (2020) Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proc Natl Acad Sci U S A 117:11029–11037. https://doi.org/10.1073/pnas.2002289117
Müller AL, de Rezende JR, Hubert CRJ, Kjeldsen KU, Lagkouvardos I, Berry D, Jorgensen BB, Loy A (2014) Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J 8:1153–1165. https://doi.org/10.1038/ismej.2013.225
Lomonosov IS (1974) Geochemistry and formation of modern hydrotherms in the Baikal Rift Zone. Nauka, Novosibirsk, p 168 (In Russian)
Verbolov VI (1996) Currents and water exchange in Lake Baikal. Water Res 23:381–391
Acknowledgements
Experimental cultivation, determination of the chemical composition of pore water in sediments and of polycyclic aromatic hydrocarbons and n-alkanes in model experiments were carried out using the equipment of the freshwater aquarium complex and Collective Instrumental Center Ultramicroanalysis at Limnological Institute SB RAS; the Sanger reaction was performed by the SB RAS Genomics Core Facility (ICBFM SB RAS, Novosibirsk, Russia).
Funding
This work was supported by State Task number 0279-2021-0006 (121032300223-1).
Author information
Authors and Affiliations
Contributions
Pavlova O.N. performed data analysis and wrote the manuscript; Izosimova O.N. and Gorshkov A.G. determined PAHs and n-alkanes in model experiments; Chernitsyna S.M. analyzed molecular biological data; Ivanov V.G. measured methane in bottom sediments and experimental samples; Pogodaeva T.V. conducted a chemical analysis of the composition of pore waters from sediments and storage cultures; Khabuev A.V. sampled sediments and described lithological core; Zemskaya T.I. coordinated the project and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Supplementary Fig. 1
Diversity of microbial community of the studied samples. The dependence of the number of phylotypes at the genus level (genetic distance 0.03) on the number of obtained sequences of the bacterial 16S rRNA gene region. Rarefaction curves of partial sequences of the bacterial 16S rRNA gene. (a) Archaea; (b) Bacteria (PNG 217 kb)
ESM 1
(DOCX 14 kb)
ESM 2
(DOCX 55 kb)
Rights and permissions
About this article
Cite this article
Pavlova, O.N., Izosimova, O.N., Chernitsyna, S.M. et al. Anaerobic oxidation of petroleum hydrocarbons in enrichment cultures from sediments of the Gorevoy Utes natural oil seep under methanogenic and sulfate-reducing conditions. Microb Ecol 83, 899–915 (2022). https://doi.org/10.1007/s00248-021-01802-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01802-y