Abstract
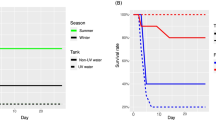
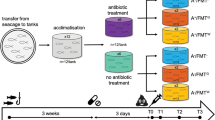
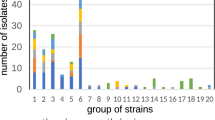
The microbiota of fish skin, the primary barrier against disease, is highly dynamic and modulated by several factors. In fish aquaculture, disease outbreaks occur mainly during early-life stages, with associated high economic losses. Antibiotic treatments sometimes remain the best option to control bacterial diseases, despite many reported negative impacts of its use on fish and associated microbiota. Notwithstanding, studies monitoring the effects of disease and antibiotic treatment on the microbiota of fingerlings are scarce. We sequenced the bacterial 16S rRNA V4 gene region using a metabarcoding approach to assess the impact of a mixed infection with Photobacterium damselae ssp. piscicida and Vibrio harveyi and subsequent antibiotic treatment with flumequine, on the skin microbiota of farmed seabass (Dicentrarchus labrax) fingerlings. Both infection and antibiotic treatment led to a significant increase in bacterial diversity and core microbial communities and impacted microbiome structure. Dysbiosis was confirmed by changes in the abundance of potential pathogenic and opportunistic bacterial taxa. Skin bacterial metabolic function was also significantly affected by flumequine administration, suggesting a detriment to fish skin health. Our results add to an increasing body of literature, showing how fish microbiome response to infection and antibiotics cannot be easily predicted.





Similar content being viewed by others
Data Availability
The raw sequences are available at NCBI Sequence Read Archive (SRA) database within the BioProject ID PRJNA741392.
References
Kelly C, Salinas I (2017) Under pressure: interactions between commensal microbiota and the teleost immune system. Front Immunol 8:1
Trivedi B (2012) Microbiome: the surface brigade. Nature 492:S60–S61
Petersen C, Round JL (2014) Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol 16:1024–1033. https://doi.org/10.1111/cmi.12308
Osadchiy V, Martin CR, Mayer EA (2019) The gut–brain axis and the microbiome: mechanisms and clinical implications. Clin Gastroenterol Hepatol 17:322–332
Legrand TPRA, Catalano SR, Wos-Oxley ML et al (2018) The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front Microbiol 8:2664. https://doi.org/10.3389/fmicb.2017.02664
Llewellyn MS, Leadbeater S, Garcia C et al (2017) Parasitism perturbs the mucosal microbiome of Atlantic Salmon. Sci Rep 7:1–10. https://doi.org/10.1038/srep43465
Rosado D, Xavier R, Severino R et al (2019) Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-55314-4
Rosado D, Pérez-Losada M, Pereira A et al (2021) Effects of aging on the skin and gill microbiota of farmed seabass and seabream. Anim Microbiome 3:10. https://doi.org/10.1186/s42523-020-00072-2
Reid KM, Patel S, Robinson AJ et al (2017) Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS One 12:e0172856. https://doi.org/10.1371/journal.pone.0172856
Borges N, Keller-Costa T, Sanches-Fernandes GMM et al (2020) Bacteriome structure, function, and probiotics in fish larviculture: the good, the bad, and the gaps. Annu Rev Anim Biosci 9:423–452. https://doi.org/10.1146/annurev-animal-062920
Parshukov AN, Kashinskaya EN, Simonov EP et al (2019) Variations of the intestinal gut microbiota of farmed rainbow trout, Oncorhynchus mykiss (Walbaum), depending on the infection status of the fish. J Appl Microbiol 127:379–395. https://doi.org/10.1111/jam.14302
Tran NT, Zhang J, Xiong F et al (2018) Altered gut microbiota associated with intestinal disease in grass carp (Ctenopharyngodon idellus). World J Microbiol Biotechnol 34:71. https://doi.org/10.1007/s11274-018-2447-2
Le Luyer J, Schull Q, Auffret P et al (2021) Dual RNAseq highlights the kinetics of skin microbiome and fish host responsiveness to bacterial infection. Anim Microbiome. https://doi.org/10.21203/rs.3.rs-108248/v1
Zhang X, Ding L, Yu Y et al (2018) The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by Ichthyophthirius multifillis. Front Immunol 9:2972. https://doi.org/10.3389/fimmu.2018.02972
Vasemägi A, Visse M, Kisand V (2017) Effect of environmental factors and an emerging parasitic disease on gut microbiome of wild salmonid fish. mSphere 2:418–435. https://doi.org/10.1128/msphere.00418-17
Miyake S, Soh M, Nursyafiq Azman M et al (2020) Insights into the microbiome of farmed Asian sea bass (Lates calcarifer) with symptoms of tenacibaculosis and description of Tenacibaculum singaporense sp. nov. Antonie Van Leeuwenhoek 113:737–752. https://doi.org/10.1007/s10482-020-01391-9
FAO (2016) FAO Fisheries & Aquaculture - Cultured Aquatic Species Information Programme - Dicentrarchus labrax (Linnaeus, 1758). http://www.fao.org/fishery/culturedspecies/Dicentrarchus_labrax/en. Accessed 12 Feb 2021
Deng Y, Zhang Y, Chen H, et al (2020) Gut–liver immune response and gut microbiota profiling reveal the pathogenic mechanisms of Vibrio harveyi in pearl gentian grouper (Epinephelus lanceolatus ♂ × E. fuscoguttatus ♀). Front Immunol 11:1. https://doi.org/10.3389/fimmu.2020.607754
Essam HM, Abdellrazeq GS, Tayel SI et al (2016) Pathogenesis of Photobacterium damselae subspecies infections in sea bass and sea bream. Microb Pathog 99:41–50. https://doi.org/10.1016/j.micpath.2016.08.003
Mohamad N, Amal MNA, Yasin ISM et al (2019) Vibriosis in cultured marine fishes: a review. Aquaculture 512:734289. https://doi.org/10.1016/j.aquaculture.2019.734289
Bakopoulos V, Volpatti D, Gusmani L et al (2003) Vaccination trials of sea bass, Dicentrarchus labrax (L.), against Photobacterium damsela subsp. piscicida, using novel vaccine mixtures. J Fish Dis 26:77–90. https://doi.org/10.1046/j.1365-2761.2003.00438.x
Almeida AR, Alves M, Domingues I, Henriques I (2019) The impact of antibiotic exposure in water and zebrafish gut microbiomes: a 16S rRNA gene-based metagenomic analysis. Ecotoxicol Environ Saf 186:109771. https://doi.org/10.1016/j.ecoenv.2019.109771
Almeida AR, Tacão M, Machado AL et al (2019) Long-term effects of oxytetracycline exposure in zebrafish: a multi-level perspective. Chemosphere 222:333–344. https://doi.org/10.1016/j.chemosphere.2019.01.147
Carlson JM, Leonard AB, Hyde ER et al (2017) Microbiome disruption and recovery in the fish Gambusia affinis following exposure to broad-spectrum antibiotic. Infect Drug Resist 10:143–154. https://doi.org/10.2147/IDR.S129055
Pindling S, Azulai D, Zheng B et al (2018) Dysbiosis and early mortality in zebrafish larvae exposed to subclinical concentrations of streptomycin. FEMS Microbiol Lett 365:188. https://doi.org/10.1093/femsle/fny188
De Liguoro M, Maraj S, Merlanti R (2019) Transgenerational toxicity of flumequine over four generations of Daphnia magna. Ecotoxicol Environ Saf 169:814–821. https://doi.org/10.1016/j.ecoenv.2018.11.077
FAO (1997) Residues of some veterinary drugs in animals and foods. Food and Nutrition Paper 14:10
Marchesi JR, Ravel J (2015) The vocabulary of microbiome research: a proposal. Microbiome 3:31. https://doi.org/10.1186/s40168-015-0094-5
Kozich JJ, Westcott SL, Baxter NT et al (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. https://doi.org/10.1371/journal.pcbi.1003531
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Lahti L, Shetty S (2018) Introduction to the microbiome R package. https://bioconductor.statistik.tu-dortmund.de/packages/3.6/bioc/vignettes/microbiome/inst/doc/vignette.html
Oksanen J, Kindt R, Legendre P et al (2008) The vegan package: community ecology package, version 1, pp 13–1. http://vegan.r-forge.r-project.org
Douglas GM, Maffei VJ, Zaneveld J, et al (2019) PICRUSt2: an improved and extensible approach for metagenome inference. bioRxiv 672295
Kanehisa M, Sato Y, Furumichi M et al (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47:D590–D595. https://doi.org/10.1093/nar/gky962
Segata N, Izard J, Waldron L et al (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. https://doi.org/10.1186/gb-2011-12-6-r60
Austin B, Austin DA (2016) Bacterial fish pathogens, 6th edn. Springer, Stirling
Joe JTX, Tseng YC, Wu JL, Lu MW (2021) The alteration of intestinal microbiota profile and immune response in Epinephelus coioides during pathogen infection. Life 11:1–18. https://doi.org/10.3390/life11020099
Laganà P, Caruso G, Minutoli E et al (2011) Susceptibility to antibiotics of Vibrio spp. and Photobacterium damsela ssp. piscicida strains isolated from Italian aquaculture farms. New Microbiol 34:53–63
Stevens J, Jackson R, Olson J (2016) Bacteria associated with lionfish (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Mar Ecol Prog Ser 558:167–180. https://doi.org/10.3354/meps11789
Wu J, Mao C, Deng Y et al (2019) Diversity and abundance of antibiotic resistance of bacteria during the seedling period in marine fish cage-culture areas of Hainan, China. Mar Pollut Bull 141:343–349. https://doi.org/10.1016/j.marpolbul.2019.02.069
López Nadal A, Peggs D, Wiegertjes GF, Brugman S (2018) Exposure to antibiotics affects saponin immersion-induced immune stimulation and shift in microbial composition in zebrafish larvae. Front Microbiol 9:2588. https://doi.org/10.3389/fmicb.2018.02588
Hooper LV, Gordon JI (2001) Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11:1R-10R. https://doi.org/10.1093/glycob/11.2.1R
Brandley BK, Schnaar RL (1986) Cell-surface carbohydrates in cell recognition and response. J Leukoc Biol 40:97–111. https://doi.org/10.1002/jlb.40.1.97
Fish KE, Collins R, Green NH et al (2015) Characterisation of the physical composition and microbial community structure of biofilms within a model full-scale drinking water distribution system. PLoS ONE 10:e0115824. https://doi.org/10.1371/journal.pone.0115824
Friedman L, Kolter R (2004) Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186:4457–4465. https://doi.org/10.1128/JB.186.14.4457-4465.2004
Brumlow CE, Luna RA, Hollister EB et al (2019) Biochemical but not compositional recovery of skin mucosal microbiome communities after disruption. Infect Drug Resist 12:399–416. https://doi.org/10.2147/IDR.S185992
Hansen M, Ingebrigtsen K, Hayton W, Horsberg T (2001) Disposition of 14C-flumequine in eel Anguilla anguilla, turbot Scophthalmus maximus and halibut Hippoglossus hippoglossus after oral and intravenous administration. Dis Aquat Organ 47:183–191. https://doi.org/10.3354/dao047183
Sohlberg S, Ingebrigtsen K, Hansen M et al (2002) Flumequine in Atlantic salmon Salmo salar: disposition in fish held in sea water versus fresh water. Dis Aquat Organ 49:39–44. https://doi.org/10.3354/dao049039
Malvisi J, Della Rocca G, Anfossi P, Giorgetti G (1997) Tissue distribution and depletion of flumequine after in-feed administration in sea bream (Sparus aurata). Aquaculture 157:197–204. https://doi.org/10.1016/S0044-8486(97)00160-9
Funding
This work was funded by the European Regional Development Fund (ERDF) through the COMPETE program and by National Funds through FCT—Foundation for Science and Technology (project PTDC/BIA-MIC/27995/2017 POCI-01–0145-FEDER-027995); DR, MP-L, and RX were supported by FCT under the Programa Operacional Potencial Humano – Quadro de Referência Estratégico Nacional funds from the European Social Fund and Portuguese Ministério da Educação e Ciência (DR doctoral grant SFRH/BD/117943/2016; MP-L: IF/00764/2013; RX: IF/00359/2015; and 2020.00854.CEECIND).
Author information
Authors and Affiliations
Contributions
DR, RX, and RS designed the research. RS collected the samples. DR performed laboratory work and analyzed the results. MP-L contributed to the statistical analysis. RX and MP-L supervised and provided intellectual content. All authors reviewed the manuscript. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
Animals in this study were reared in a commercial fish farm located in the estuarine environment of the Alvor Estuary (Portimão), southern Portugal. Fish were handled by the fish farm employees and samples were taken non-invasively. According to the Portuguese legislation DL Nº 113/2013, our work does not involve animal experimentation and therefore is exempted from the need of ethical approval.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rosado, D., Pérez-Losada, M., Severino, R. et al. Monitoring Infection and Antibiotic Treatment in the Skin Microbiota of Farmed European Seabass (Dicentrarchus Labrax) Fingerlings. Microb Ecol 83, 789–797 (2022). https://doi.org/10.1007/s00248-021-01795-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01795-8