Abstract
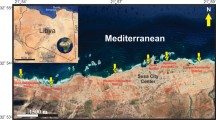
Seamounts are often covered with Fe and Mn oxides, known as ferromanganese (Fe–Mn) crusts. Future mining of these crusts is predicted to have significant effects on biodiversity in mined areas. Although microorganisms have been reported on Fe–Mn crusts, little is known about the role of crusts in shaping microbial communities. Here, we investigated microbial communities based on 16S rRNA gene sequences retrieved from Fe–Mn crusts, coral skeleton, calcarenite, and biofilm at crusts of the Rio Grande Rise (RGR). RGR is a prominent topographic feature in the deep southwestern Atlantic Ocean with Fe–Mn crusts. Our results revealed that crust field of the RGR harbors a usual deep-sea microbiome. No differences were observed on microbial community diversity among Fe–Mn substrates. Bacterial and archaeal groups related to oxidation of nitrogen compounds, such as Nitrospirae, Nitrospinae phyla, Candidatus Nitrosopumilus within Thaumarchaeota group, were present on those substrates. Additionally, we detected abundant assemblages belonging to methane oxidation, i.e., Methylomirabilales (NC10) and SAR324 (Deltaproteobacteria). The chemolithoautotrophs associated with ammonia-oxidizing archaea and nitrite-oxidizing bacteria potentially play an important role as primary producers in the Fe–Mn substrates from RGR. These results provide the first insights into the microbial diversity and potential ecological processes in Fe–Mn substrates from the Atlantic Ocean. This may also support draft regulations for deep-sea mining in the region.






Similar content being viewed by others
Data Availability
The sequencing reads generated for this study can be found in the National Centre for Biotechnology Information (NCBI) database under the BioProject PRJNA638744.
References
Benites M, Hein J, Mizell K, Blackburn T, Jovane L (2020) Genesis and evolution of ferromanganese crusts from the summit of Rio Grande Rise, Southwest Atlantic Ocean. Minerals 10:349. https://doi.org/10.3390/min10040349
Blothe MA, Wegorzewski C, Muller FS, Kuhn T, Schippers A (2015) Manganese-cycling microbial communities inside deep-sea manganese nodules. Environ Sci Technol 49:7692–7700. https://doi.org/10.1021/es504930v
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Burnett BR, Nealson KH (1981) Organic films and microorganisms associated with manganese nodules. Deep-Sea Res I Oceanogr Res Pap 28:637–645
Cáceres MD, Legendre P, Moretti M (2010) Improving indicator species analysis by combining groups of sites. Oikos 119:1674–1684. https://doi.org/10.1111/j.1600-0706.2010.18334.x
Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Cavalcanti JAD, Santos RV, Lacasse CM, Rojas JNL, Nobrega M (2015) Potential mineral resources of phosphates and trace elements on the Rio Grande Rise, South Atlantic Ocean. Nearshore underwater mining: critical commodities for the future UMC, Tampa Bay
Du Z, Wang Z, Zhao J-X, Chen G (2016) Woeseia oceani gen. nov., sp. nov., a novel chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov. Int J Syst Evol Microbiol 66. https://doi.org/10.1099/ijsem.0.000683
Etnoyer PJ, Wood J, Shirley TC (2010) How large is the seamount biome. Oceanography 23:206–209. https://doi.org/10.5670/oceanog.2010.96
Hein JR, Koschinsky A (2014) Deep-ocean ferromanganese crusts and nodules. In: Holland HD, Turekian KK (eds) Treatise on geochemistry 2nd ed. Elsevier, pp 273–291. https://doi.org/10.1016/B978-0-08-095975-7.01111-6
Hoffmann K, Bienhold C, Buttigieg PL, Knittel K, Laso-Perez R, Rapp JZ, Boetius A, Offre P (2020) Diversity and metabolism of Woeseiales bacteria, global members of marine sediment communities. ISME J 14:1042–1056. https://doi.org/10.1038/s41396-020-0588-4
Huo Y, Cheng H, Anton F, Wang C, Jiang X, Pan J, Wu M, Xu X (2015) Ecological functions of uncultured microorganisms in the cobalt-rich ferromanganese crust of a seamount in the central Pacific are elucidated by fosmid sequencing. Acta Oceanol Sin 34:92–113. https://doi.org/10.1007/s13131-015-0650-7
Jianga X-D, Suna X-M, Guana Y (2017) Biogenic mineralization in the ferromanganese nodules and crusts from the South China Sea. J Asia Earth Sci 171:46–59. https://doi.org/10.1016/j.jseaes.2017.07.050
Jørgensen BB, Boetius A (2007) Feast and famine—microbial life in the deep-sea bed. Nat Rev Microbiol 5:770–781. https://doi.org/10.1038/nrmicro1745
Kato S, Okumura T, Uematsu K, Hirai M, Iijima K, Usui A, Suzuki K (2018) Heterogeneity of microbial communities on deep-sea ferromanganese crusts in the Takuyo-Daigo seamount. Microbes Environ 33:366–377. https://doi.org/10.1264/jsme2.ME18090
Kato S, Hirai M, Ohkuma M, Suzuki K (2019) Microbial metabolisms in an abyssal ferromanganese crust from the Takuyo-Daigo Seamount as revealed by metagenomics. PLoS ONE 14:e0224888. https://doi.org/10.1371/journal.pone.0224888
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software ver. 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kellogg CA (2019) Microbiomes of stony and soft deep-sea corals share rare core bacteria. Microbiome 7:90. https://doi.org/10.1186/s40168-019-0697-3
Levin LA, Mengerink K, Gjerde KM, Rowden AA, Van Dover CL, Clark MR, Ramirez-Llodra E, Currie B et al (2016) Defining “serious harm” to the marine environment in the context of deep seabed mining. Mar Policy 74(245-259):2016–2259. https://doi.org/10.1016/j.marpol.2016.09.032
Li J, Mara P, Schubotz F, Sylvan JB, Burgaud G, Klein F, Beaudoin D, Wee SY, Dick HJB, Lott S, Cox R, Meyer LAE, Quémener M, Blackman DK, Edgcomb VP (2020) Recycling and metabolic flexibility dictate life in the lower oceanic crust. Nature 579:250–255. https://doi.org/10.1038/s41586-020-2075-5
Lindh MV, Maillot BM, Shulse CN, Gooday AJ, Amon DJ, Smith CR, Church MJ (2017) From the surface to the deep-sea: bacterial distributions across polymetallic nodule fields in the clarion-clipperton zone of the Pacific Ocean. Front Microbiol 8:1–12. https://doi.org/10.3389/fmicb.2017.01696
Liu Q, Huo YY, Wu Y-H, Bai Y, Yuan Y, Xu D, Wang J, Wang C-S, Xu X-W (2019) Bacterial community on a Guyot in the Northwest Pacific Ocean influenced by physical dynamics and environmental variables. J Geophys Res Biogeosci 124:2883–2897. https://doi.org/10.1029/2019JG005066
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Sci 353:1272–1277. https://doi.org/10.1126/science.aaf4507
Love MI, Huber W, Andres S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mcmurdie PJ, Holmes S (2012) Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput:235–246. https://doi.org/10.1142/9789814366496_0023
Mohriak WU, Nobrega M, Odegard ME, Gomes BS, Dickson W (2010) Geological and geophysical interpretation of the Rio Grande Rise, south-eastern Brazilian margin: extensional tectonics and rifting of continental and oceanic crusts. Pet Geosci 16:231–245. https://doi.org/10.1144/1354-079309-910
Molari M, Janssen F, Vonnahme T, Boetius A (2020) Microbial communities associated with sediments and polymetallic nodules of the Peru Basin. Biogeosci Discuss., in review. https://doi.org/10.5194/bg-2020-11
Montserrat F, Guilhon M, Correa PVF, Bergo NM, Signori CN, Tura PM, Maly M de los S et al (2019) Deep-sea mining on the Rio Grande Rise (Southwestern Atlantic): review on environmental baseline, ecosystem services and potential impacts. Deep-Sea Res I Oceanogr Res Pap 145:31–58. https://doi.org/10.1016/j.dsr.2018.12.007
Mußmann M, Pjevac P, Kruger K, Dyksma S (2017) Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J 11:1276–1281. https://doi.org/10.1038/ismej.2016.185
Nitahara S, Kato S, Urabe T, Usui A, Yamagishi A (2011) Molecular characterization of the microbial community in hydrogenetic ferromanganese crusts of the Takuyo-Daigo Seamount, northwest Pacific. FEMS Microbiol Lett 321:121–129. https://doi.org/10.1111/j.1574-6968.2011.02323.x
Nitahara S, Kato S, Usui A, Urabe T, Suzuki K, Yamagishi A (2017) Archaeal and bacterial communities in deep- sea hydrogenetic ferromanganese crusts on old seamounts of the northwestern Pacific. PLoS ONE 12:e0173071. https://doi.org/10.1371/journal.pone.0173071
Oksanen, J, Blanchet FG, Kindt R, Legendre P, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: community ecology package. R package version 1.17-2
Orcutt BN, Bradley JA, Brazelton WJ, Estes ER, Goordial JM, Huber JA, Jones RM, Mahmoudi N, Marlow JJ, Murdock S, Pachiadaki M (2020) Impacts of deep-sea mining on microbial ecosystem services. Limnol Oceanogr 9999:1–2. https://doi.org/10.1002/lno.11403
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Queiroz LL, Bendia AG, Duarte RTD, Gracas DA, Silva ALC, Nakayama CR, Sumida PY, Lima AOS, Negano Y, Fujikura K, Kitazato H, Pellizari VH (2020) Bacterial diversity in deep-sea sediments under influence of asphalt seep at the Sao Paulo Plateau. Antonie Van Leeuwenhoek 113(5):707–717. https://doi.org/10.1007/s10482-020-01384-8
Shank TM (2010) Seamounts: deep-ocean laboratories of faunal connectivity, evolution and endemism. Oceanogr 23:108–122. https://doi.org/10.5670/oceanog.2010.65
Sheik CS, Jain S, Dick GJ (2014) Metabolic flexibility of enigmatic SAR 324 revealed through metagenomics and metatranscriptomics. Environ Microbiol 16:304–317. https://doi.org/10.1111/1462-2920.12165
Sheik CS, Reese BK, Twing KI, Sylvan JB, Grim SL, Schrenk MO, Sogin ML, Colwell FS (2018) Identification and removal of contaminant sequences from ribosomal gene databases: lessons from the census of deep life. Front Microbiol 8. https://doi.org/10.3389/fmicb.2018.00840
Shiraishi F, Mitsunobu S, Suzuki K, Hoshino T, Morono Y, Inagaki F (2016) Dense microbial community on a ferromanganese nodule from the ultra-oligotrophic South Pacific gyre: Implications for biogeochemical cycles. Earth Planet Sci Lett 447:10–20
Shulse C, Maillot B, Smith CR, Church MJ (2016) Polymetallic nodules, sediments, and deep waters in the equatorial North Pacific exhibit highly diverse and distinct bacterial, archaeal, and microeukaryotic communities. Microbiol Open 6:e00428. https://doi.org/10.1002/mbo3.428
Sunda WG, Kieber DJ (1994) Oxidation of humic substances by manganese oxides yields low-molecular-weight organic substrates. Nature 367:62–64. https://doi.org/10.1038/367062a0
Templeton AS, Knowles EJ, Eldridge DL, Arey BW, Dohnalkova AC, Webb SM, Bailey BE, Tebo BM, Staudigel H (2009) A seafloor microbial biome hosted within incipient ferromanganese crusts. Nat Geosci 2:872–876. https://doi.org/10.1038/ngeo696
Usui A, Nishi K, Sato H, Nakasato Y, Thornton B, Kashiwabara T, Tokumaru A, Sakaguchi A, Yamaoka K, Kato S, Nitahara S, Suzuki K, Iijima K, Urabe T (2017) Continuous growth of hydro- genetic ferromanganese crusts since 17myr ago on Takuyo-Daigo Seamount, NW Pacific, at water depths of 800–5500 m. Ore Geol Rev 87:71–87. https://doi.org/10.1016/j.oregeorev.2016.09.032
Versantvoort W, Guerrero-Cruz S, Speth DR, Frank J, Gambelli L, Cremers G, Van Alen T, Jetten MSM, Kartal K, Op den Camp HJM, Reimann J (2019) Comparative genomics of Candidatus Methylomirabilis species and description of Ca. Methylomirabilis Lanthanidiphila. Front Microbiol 9:1672. https://doi.org/10.3389/fmicb.2018.01672
Wang X, Muller WEG (2009) Marine biominerals: perspectives and challenges for polymetallic nodules and crusts. Trends Biotechnol 27:375–383. https://doi.org/10.1016/j.tibtech.2009.03.004
Wang X, Schlossmacher U, Wiens M, Schroeder HC, Muller WEG (2009) Biogenic origin of polymetallic nodules from the Clarion- Clipperton Zone in the Eastern Pacific Ocean: electron microscopic and EDX evidence. Mar Biotechnol 11:99–108
Wessel P, Sandwell DT, Kim S-S (2010) The global seamount census. Oceanogr 23:24–33. https://doi.org/10.5670/oceanog.2010.60
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer Publishing Company, Incorporated, New York
Wu YH, Liao L, Wang CS, Ma WL, Meng FX, Wu M, Xu XW (2013) A comparison of microbial communities in deep-sea polymetallic nodules and the surrounding sediments in the Pacific Ocean. Deep-Sea Res I Oceanogr Res Pap 79:40–49. https://doi.org/10.1016/j.dsr.2013.05.004
Yang S-H, Tang S-L (2019) Endolithic microbes in coral skeletons: algae or bacteria? In: Zhiyong L (ed) Symbiotic Microbiomes of Coral Reefs Sponges and Corals, Springer, Dordrecht, pp 43-53. DOI. https://doi.org/10.1007/978-94-024-1612-1_4
Yu H, Leadbetter JR (2020) Bacterial chemolithoautotrophy via manganese oxidation. Nature 583:453–458. https://doi.org/10.1038/s41586-020-2468-5
Acknowledgments
We thank the captain and the crew of the Royal Research Ship Discovery cruise DY094 for their data and sampling support, as well as LECOM’s research team and Rosa C. Gamba for their scientific support. Also, we thank Mariana Benites and Pedro M. Tura for all sampling and scientific support, and all MarineE-tech members.
Funding
This study was funded by the São Paulo Research Foundation (FAPESP), Grant number: 14/50820-7, Project Marine ferromanganese deposits: a major resource of E-Tech elements, which is an international collaboration between Natural Environment Research Council (NERC, UK) and FAPESP (BRA). NMB thanks the Ph.D. scholarship financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Supplementary Information
ESM 1
(DOCX 207 kb)
Rights and permissions
About this article
Cite this article
Bergo, N.M., Bendia, A.G., Ferreira, J.C.N. et al. Microbial Diversity of Deep-Sea Ferromanganese Crust Field in the Rio Grande Rise, Southwestern Atlantic Ocean. Microb Ecol 82, 344–355 (2021). https://doi.org/10.1007/s00248-020-01670-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01670-y