Abstract
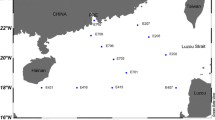
Recently, an increasing number of studies have focused on the biogeographic distribution of marine microorganisms. However, the extent to which geographic distance can affect marine microbial communities is still unclear, especially for the microbial communities in well-connected surface seawaters. In this study, the bacterial community compositions of 21 surface seawater samples, that were distributed over a distance of 7800 km, were surveyed to investigate how bacterial community similarity changes with increasing geographical distance. Proteobacteria and Bacteroidetes were the dominant bacterial phyla, with Proteobacteria accounting for 52.6–92.5% and Bacteroidetes comprising 3.5–46.9% of the bacterial communities. A significant bacterial distance-decay relationship was observed in the well-connected Southern Ocean surface seawater. The number of pairwise shared operational taxonomic units (OTUs), and community similarities tended to decrease with increasing geographic distance. Calculation of the similarity indices with all, abundant or rare OTUs did not affect the observed distance-decay relationship. Spatial distance can largely explain the observed bacterial community variation. This study shows that even in well-connected surface waters, bacterial distance-decay patterns can be found as long as the geographical distance is great enough. The biogeographic patterns should then be present for marine microorganisms considering the large size and complexity of the marine ecosystem.






Similar content being viewed by others
References
Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. https://doi.org/10.1038/nrmicro1341
Zhang G, Li Q, Sun S (2018) Diversity and distribution of parasitic angiosperms in China. Ecol Evol 8:4378–4386. https://doi.org/10.1002/ece3.3992
Valdecasas AG, Camacho AI, Pelaez ML (2006) Do small animals have a biogeography? Exp Appl Acarol 40:133–144. https://doi.org/10.1007/s10493-006-9030-5
Neill WT (1964) Biogeography: the distribution of animals and plants. BSCS pamphlets 18:1–36
Hernando-Morales V, Ameneiro J, Teira E (2017) Water mass mixing shapes bacterial biogeography in a highly hydrodynamic region of the Southern Ocean. Environ Microbiol 19:1017–1029. https://doi.org/10.1111/1462-2920.13538
Wilkins D, van Sebille E, Rintoul SR, Lauro FM, Cavicchioli R (2013) Advection shapes Southern Ocean microbial assemblages independent of distance and environment effects. Nat Commun. 4:2457. https://doi.org/10.1038/ncomms3457
Pinel-Galzi A, Traore O, Sere Y, Hebrard E, Fargette D (2015) The biogeography of viral emergence: rice yellow mottle virus as a case study. Curr Opin Virol 10:7–13. https://doi.org/10.1016/j.coviro.2014.12.002
Giovannoni SJ, Vergin KL (2012) Seasonality in ocean microbial communities. Science 335:671–676. https://doi.org/10.1126/science.1198078
Ghiglione JF, Galand PE, Pommier T, Pedros-Alio C, Maas EW, Bakker K, Bertilson S, Kirchmanj DL, Lovejoy C, Yager PL, Murray AE (2012) Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci U S A 109:17633–17638. https://doi.org/10.1073/pnas.1208160109
Li WL, Huang JM, Zhang PW, Cui GJ, Wei ZF, Wu YZ, Gao ZM, Han Z, Wang Y (2018) Periodic and spatial spreading of alkanes and Alcanivorax bacteria in deep waters of the Mariana trench. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02089-18
de Wit R, Bouvier T (2006) 'Everything is everywhere, but, the environment selects'; what did Baas Becking and Beijerinck really say? Environ Microbiol 8:755–758. https://doi.org/10.1111/j.1462-2920.2006.01017.x
Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA (2013) Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci U S A 110:4651–4655. https://doi.org/10.1073/pnas.1217767110
Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA (2012) The Western English Channel contains a persistent microbial seed bank. ISME J 6:1089–1093. https://doi.org/10.1038/ismej.2011.162
Tfouni LV, Klatzky RL (1983) A discourse analysis of deixis: pragmatic, cognitive and semantic factors in the comprehension of 'this', 'that', 'here' and 'there'. J Child Lang 10:123–133
Li Y, Sun LL, Sun ML, Su HN, Zhang XY, Xie BB, Chen XL, Zhang YZ, Qin QL (2018) Vertical and horizontal biogeographic patterns and major factors affecting bacterial communities in the open South China Sea. Sci Rep 8:8800. https://doi.org/10.1038/s41598-018-27191-w
Agogue H, Lamy D, Neal PR, Sogin ML, Herndl GJ (2011) Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20:258–274. https://doi.org/10.1111/j.1365-294X.2010.04932.x
Smith PJ, Steinke D, McMillan PJ, Stewart AL, McVeagh SM, Diaz de Astarloa JM, Welsford D, Ward RD (2011) DNA barcoding highlights a cryptic species of grenadier Macrourus in the Southern Ocean. J Fish Biol 78:355–365. https://doi.org/10.1111/j.1095-8649.2010.02846.x
Zakrzewski M, Goesmann A, Jaenicke S, Junemann S, Eikmeyer F, Szczepanowski R, Al-Soud WA, Sorensen S, Puhler A, Schluter A (2012) Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J Biotechnol 158:248–258. https://doi.org/10.1016/j.jbiotec.2012.01.020
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Mo Y, Zhang W, Yang J, Lin Y, Yu Z, Lin S (2018) Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J 12:2198–2210. https://doi.org/10.1038/s41396-018-0153-6
Li HB, Zeng J, Ren LJ, Wang JJ, Xing P, Wu QLL (2017) Contrasting patterns of diversity of abundant and rare bacterioplankton in freshwater lakes along an elevation gradient. Limnol Oceanogr 62:1570–1585. https://doi.org/10.1002/lno.10518
Clarke KRGR (2015) PRIMERv7: user manual/tutorial. PRIMER-E Ltd., Plymouth
Ter Braak CJF, Smilauer P (2012) Canoco reference manual and user's guide: software for ordination (version 5.0). Microcomputer power. Ithace, New York
Dray S, Legendre P, Peres-Neto PR (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196:483–493. https://doi.org/10.1016/j.ecolmodel.2006.02.015
Whitton BA, Potts M (2013) The ecology of cyanobacteria. Springer Netherlands
Vincent WF (2000) Cyanobacterial dominance in the polar regions. Springer Netherlands
Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. https://doi.org/10.1128/MMBR.00051-12
Zhang ZZ, Lu Y, Hsu HT (2007) Detecting surface geostrophic currents using wavelet filter from satellite geodesy. Sci China Ser D 50:918–926
Ferrari R, Wunsch C (2009) Ocean circulation kinetic energy: reservoirs, sources, and sinks. Annu Rev Fluid Mech 41:253–282
Zinger L, Boetius A, Ramette A (2014) Bacterial taxa-area and distance-decay relationships in marine environments. Mol Ecol 23:954–964. https://doi.org/10.1111/mec.12640
Zeglin LH (2015) Stream microbial diversity in response to environmental changes: review and synthesis of existing research. Front Microbiol 6:454. https://doi.org/10.3389/fmicb.2015.00454
Yu SX, Pang YL, Wang YC, Li JL, Qin S (2018) Distribution of bacterial communities along the spatial and environmental gradients from Bohai Sea to northern Yellow Sea. Peer J 6:e4272. https://doi.org/10.7717/peerj.4272
Villarino E, Watson JR, Jonsson B, Gasol JM, Salazar G, Acinas SG, Estrada M, Massana R, Logares R, Giner CR, Pernice MC, Olivar MP, Citores L, Corell J, Rodriguez-Ezpeleta N, Acuna JL, Molina-Ramirez A, Gonzalez-Gordillo JI, Cozar A, Marti E, Cuesta JA, Agusti S, Fraile-Nuez E, Duarte CM, Irigoien X, Chust G (2018) Large-Scale Ocean connectivity and planktonic body size. Nat Commun 9:142. https://doi.org/10.1038/s41467-017-02535-8
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB (2012) Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. https://doi.org/10.1038/nrmicro2795
Yang J, Jiang H, Wu G, Liu W, Zhang G (2016) Distinct factors shape aquatic and sedimentary microbial community structures in the lakes of Western China. Front Microbiol 7:1782. https://doi.org/10.3389/fmicb.2016.01782
Funding
The work was supported by National Key R&D Program of China (grants 2018YFC1406700, 2018YFC1406702, 2018YFC1406704, and 2018YFC1406706), the National Science Foundation of China (grants 31670063, 31670038, U1706207, 31800107, 91851205, and 31870101), AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (2017ASTCP-OS14), Taishan Scholars Program of Shandong Province (tspd20181203), the Key Research Project of Shandong Province (2017GSF21106), and the Young Scholars Program of Shandong University (2016WLJH36).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, ZB., Sun, YY., Li, Y. et al. Significant Bacterial Distance-Decay Relationship in Continuous, Well-Connected Southern Ocean Surface Water. Microb Ecol 80, 73–80 (2020). https://doi.org/10.1007/s00248-019-01472-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01472-x