Abstract
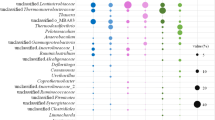
Butyrate is one of the most important intermediates during anaerobic digestion of protein wastewater, and its oxidization is considered as a rate-limiting step during methane production. However, information on syntrophic butyrate-oxidizing bacteria (SBOB) is limited due to the difficulty in isolation of pure cultures. In this study, two anaerobic chemostats fed with butyrate as the sole carbon source were operated at different dilution rates (0.01/day and 0.05/day). Butyrate- and acetate-oxidizing bacteria in both chemostats were investigated, combining DNA-Stable Isotope Probing (DNA-SIP) and 16S rRNA gene high-throughput sequencing. The results showed that, in addition to known SBOB, Syntrophomonas, other species of unclassified Syntrophomonadaceae were putative butyrate-oxidizing bacteria. Species of Mesotoga, Aminivibrio, Acetivibrio, Desulfovibrio, Petrimonas, Sedimentibacter, unclassified Anaerolineae, unclassified Synergistaceae, unclassified Spirochaetaceae, and unclassified bacteria may contribute to acetate oxidation from butyrate metabolism. Among them, the ability of butyrate oxidation was unclear for species of Sedimentibacter, unclassified Synergistaceae, unclassified Spirochaetaceae, and unclassified bacteria. These results suggested that more unknown species participated in the degradation of butyrate. However, the corresponding function and pathway for butyrate or acetate oxidization of these labeled species need to be further investigated.






Similar content being viewed by others
References
Ramsay IR, Pullammanappallil PC (2001) Protein degradation during anaerobic wastewater treatment: derivation of stoichiometry. Biodegradation 12:247–256. https://doi.org/10.1023/a:1013116728817
Batstone DJ, Pind PF, Angelidaki I (2003) Kinetics of thermophilic, anaerobic oxidation of straight and branched chain butyrate and valerate. Biotechnol Bioeng 84:195–204. https://doi.org/10.1002/bit.10753
Tang Y, Shigematsu T, Morimura S, Kida K (2005) Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J Biosci Bioeng 99:150–164. https://doi.org/10.1263/jbb.99.150
Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280. https://doi.org/10.1016/j.ijpharm.2004.07.010
Schink B, Stams AJM (2013) Syntrophism among prokaryotes. In: Rosenberg E, Delong E, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, New York, pp 471–493
Mcinerney MJ, Bryant MP, Hespell RB, Costerton JW (1981) Syntrophomonas wolfei gen. Nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol 41:1029–1039
Wu C, Liu X, Dong X (2006) Syntrophomonas cellicola sp. nov., a spore-forming syntrophic bacterium isolated from a distilled-spirit-fermenting cellar, and assignment of Syntrophospora bryantii to Syntrophomonas bryantii comb. nov. Int J Syst Evol Microbiol 56:2331–2335. https://doi.org/10.1099/ijs.0.64377-0
Lorowitz WH, Zhao H, Bryant MP (1989) Syntrophomonas wolfei subsp. saponavida subsp. nov., a long-chain fatty-acid-degrading, anaerobic, syntrophic bacterium; Syntrophomonas wolfei subsp. wolfei subsp. nov.; and emended descriptions of the genus and species. Int J Syst Bacteriol 39:122–126. https://doi.org/10.1099/00207713-39-2-122
Wu C, Dong X, Liu X (2007) Syntrophomonas wolfei subsp. methylbutyratica subsp. nov., and assignment of Syntrophomonas wolfei subsp. saponavida to Syntrophomonas saponavida sp. nov. comb. nov. Syst Appl Microbiol 30:376–380. https://doi.org/10.1016/j.syapm.2006.12.001
Wu C, Liu X, Dong X (2006) Syntrophomonas erecta subsp. sporosyntropha subsp. nov., a spore-forming bacterium that degrades short chain fatty acids in co-culture with methanogens. Syst Appl Microbiol 29:457–462. https://doi.org/10.1016/j.syapm.2006.01.003
Zhang C, Liu X, Dong X (2005) Syntrophomonas erecta sp. nov., a novel anaerobe that syntrophically degrades short-chain fatty acids. Int J Syst Evol Microbiol 55:799–803. https://doi.org/10.1099/ijs.0.63372-0
Zhao HX, Yang DC, Woese CR, Bryant MP (1990) Assignment of Clostridium bryantii to Syntrophospora bryantii gen. Nov., comb. nov. on the basis of a 16S rRNA sequence analysis of its crotonate-grown pure culture. Int J Syst Bacteriol 40:40–44. https://doi.org/10.1099/00207713-40-1-40
Zhang C, Liu X, Dong X (2004) Syntrophomonas curvata sp. nov., an anaerobe that degrades fatty acids in co-culture with methanogens. Int J Syst Evol Microbiol 54:969–973. https://doi.org/10.1099/ijs.0.02903-0
Sousa DZ, Smidt H, Alves MM, Stams AJ (2007) Syntrophomonas zehnderi sp. nov., an anaerobe that degrades long-chain fatty acids in co-culture with Methanobacterium formicicum. Int J Syst Evol Microbiol 57:609–615. https://doi.org/10.1099/ijs.0.64734-0
Hatamoto M, Imachi H, Fukayo S, Ohashi A, Harada H (2007) Syntrophomonas palmitatica sp. nov., an anaerobic, syntrophic, long-chain fatty-acid-oxidizing bacterium isolated from methanogenic sludge. Int J Syst Evol Microbiol 57:2137–2142. https://doi.org/10.1099/ijs.0.64981-0
Svetlitshnyi V, Rainey F, Wiegel J (1996) Thermosyntropha lipolytica gen. Nov., sp. nov., a lipolytic, anaerobic, alkalitolerant, thermophilic bacterium utilizing short- and long-chain fatty acids in syntrophic coculture with a methanogenic archaeum. Int J Syst Bacteriol 46:1131–1137. https://doi.org/10.1099/00207713-46-4-1131
Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H (2000) Syntrophothermus lipocalidus gen. Nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int J Syst Evol Microbiol 50(Pt 2):771–779. https://doi.org/10.1099/00207713-50-2-771
Jackson BE, Bhupathiraju VK, Tanner RS, Woese CR, Mcinerney MJ (1999) Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol 171:107–114. https://doi.org/10.1007/s002030050685
Kendall M, Liu Y (2006) Butyrate- and propionate-degrading syntrophs from permanently cold marine sediments in Skan Bay, Alaska, and description of Algorimarina butyrica gen. Nov., sp nov. FEMS Microbiol Lett 262:107–114. https://doi.org/10.1111/j.1574-6968.2006.00380.x
Mcinerney MJ, Rohlin L, Mouttaki H, Kim UM, Krupp RS, Rios-Hernandez L, Sieber J, Struchtemeyer CG, Bhattacharyya A, Campbell JW (2007) The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci U S A 104:7600–7605. https://doi.org/10.1073/pnas.0610456104
Sieber JR, Sims DR, Han C, Kim E, Lykidis A, Lapidus AL, Mcdonnald E, Rohlin L, Culley DE, Gunsalus R (2010) The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production. Environ Microbiol 12:2289–2301. https://doi.org/10.1111/j.1462-2920.2010.02237.x
Hansen KH, Ahring BK, Raskin L (1999) Quantification of syntrophic fatty acid-β-oxidizing bacteria in a mesophilic biogas reactor by oligonucleotide probe hybridization. Appl Environ Microbiol 65:4767–4774. https://doi.org/10.1016/j.ydbio.2005.05.019
Menes RJ, Travers D (2006) Detection of fatty acid beta-oxidizing syntrophic bacteria by fluorescence in situ hybridization. Water Sci Technol 54:33–39. https://doi.org/10.2166/wst.2006.483
Zellner G, Macario AJL, Macario ECD (1997) A study of three anaerobic methanogenic bioreactors reveals that syntrophs are diverse and different from reference organisms 1. FEMS Microbiol Eco 22:295–301. https://doi.org/10.1111/j.1574-6941.1997.tb00381.x
Dumont MG, Murrell JC (2005) Stable isotope probing - linking microbial identity to function. Nat Rev Microbiol 3:499–504. https://doi.org/10.1038/nrmicro1162
Chauhan A, Ogram A (2006) Fatty acid-oxidizing consortia along a nutrient gradient in the Florida Everglades. Appl Environ Microbiol 72:2400–2406. https://doi.org/10.1128/AEM.72.4.2400-2406.2006
Hatamoto M, Imachi H, Yashiro Y, Ohashi A, Harada H (2008) Detection of active butyrate-degrading microorganisms in methanogenic sludges by RNA-based stable isotope probing. Appl Environ Microbiol 74:3610–3614. https://doi.org/10.1128/AEM.00045-08
Kristiansen A, Lindholst S, Feilberg A, Nielsen PH, Neufeld JD, Nielsen JL (2011) Butyric acid- and dimethyl disulfide-assimilating microorganisms in a biofilter treating air emissions from a livestock facility. Appl Environ Microbiol 77:8595–8604. https://doi.org/10.1128/AEM.06175-11
Liu P, Qiu Q, Lu Y (2011) Syntrophomonadaceae-affiliated species as active butyrate-utilizing syntrophs in paddy field soil. Appl Environ Microbiol 77:3884–3887. https://doi.org/10.1128/AEM.00190-11
Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y (2006) Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl Environ Microbiol 72:1623–1630. https://doi.org/10.1128/AEM.72.2.1623-1630.2006
Tang YQ, Shigematsu T, Morimura S, Kida K (2007) Effect of dilution rate on the microbial structure of a mesophilic butyrate-degrading methanogenic community during continuous cultivation. Appl Microbiol Biotechnol 75:451–465. https://doi.org/10.1007/s00253-006-0819-2
Jiang X, Hayashi J, Sun ZY, Yang L, Tang YQ, Oshibe H, Osaka N, Kida K (2013) Improving biogas production from protein-rich distillery wastewater by decreasing ammonia inhibition. Process Biochem 48:1778–1784. https://doi.org/10.1016/j.procbio.2013.08.014
Shigematsu T, Tang Y, Kawaguchi H, Ninomiya K, Kijima J, Kobayashi T, Morimura S, Kida K (2003) Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J Biosci Bioeng 96:547–558. https://doi.org/10.1016/s1389-1723(04)70148-6
Lueders T, Manefield M, Friedrich MW (2010) Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol 6:73–78. https://doi.org/10.1046/j.1462-2920.2003.00536.x
Neufeld JD, Schäfer H, Cox MJ, Boden R, Mcdonald IR, Murrell JC (2007) Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol andmethylamine metabolism. ISME J 1:480–491. https://doi.org/10.1038/ismej.2007.65
Shigematsu T, Tang Y, Mizuno Y, Kawaguchi H, Morimura S, Kida K (2006) Microbial diversity of mesophilic methanogenic consortium that can degrade long-chain fatty acids in chemostat cultivation. J Biosci Bioeng 102:535–544. https://doi.org/10.1263/jbb.102.535
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072. https://doi.org/10.1128/AEM.66.11.5066-5072.2000
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T, Liu WT (2015) Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9:1710–1722. https://doi.org/10.1038/ismej.2014.256
Xia Y, Wang Y, Wang Y, Chin FYL, Zhang T (2016) Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol Biofuels 9(111):111. https://doi.org/10.1186/s13068-016-0524-z
Wang G, Li Q, Gao X, Wang XC (2018) Synergetic promotion of syntrophic methane production from anaerobic digestion of complex organic wastes by biochar: performance and associated mechanisms. Bioresour Technol 250:812–820. https://doi.org/10.1016/j.biortech.2017.12.004
Ben AGZ, Thioye A, Cayol JL, Joseph M, Fauque G, Labat M (2018) Characterization of Desulfovibrio salinus sp. nov., a slightly halophilic sulfate-reducing bacterium isolated from a saline lake in Tunisia. Int J Syst Evol Microbiol 68:715–720. https://doi.org/10.1099/ijsem.0.002567
Daisuke S, Atsuko U, Toshiko S, Yoshimi O, Katsuji U (2010) Desulfovibrio butyratiphilus sp. nov., a gram-negative, butyrate-oxidizing, sulfate-reducing bacterium isolated from an anaerobic municipal sewage sludge digester. Int J Syst Evol Microbiol 60:595–602. https://doi.org/10.1099/ijs.0.013771-0
Na H, Lever MA, Kjeldsen KU, Schulz F, Jørgensen BB (2015) Uncultured Desulfobacteraceae and Crenarchaeotal group C3 incorporate (13) C-acetate in coastal marine sediment. Environ Microbiol Rep 7:614–622. https://doi.org/10.1111/1758-2229.12296
Grabowski A, Tindall BJ, Bardin V, Blanchet D, Jeanthon C (2005) Petrimonas sulfuriphila gen. Nov., sp. nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int J Syst Evol Microbiol 55:1113–1121. https://doi.org/10.1099/ijs.0.63426-0
Imachi H, Sakai S, Kubota T, Miyazaki M, Saito Y, Takai K (2016) Sedimentibacter acidaminivorans sp. nov., an anaerobic, amino acids-utilizing bacterium isolated from marine subsurface sediment. Int J Syst Evol Microbiol 66:1293–1300. https://doi.org/10.1099/ijsem.0.000878
Obst M, Krug A, Luftmann H, Steinbüchel A (2005) Degradation of cyanophycin by Sedimentibacter hongkongensis strain KI and Citrobacter amalonaticus strain G isolated from an anaerobic bacterial consortium. Appl Environ Microbiol 71:3642–3652. https://doi.org/10.1128/AEM.71.7.3642-3652.2005
Hao LT, Zhang BG, Cheng M, Feng CP (2016) Effects of various organic carbon sources on simultaneous V(V) reduction and bioelectricity generation in single chamber microbial fuel cells. Bioresour Technol 201:105–110. https://doi.org/10.1016/j.biortech.2015.11.060
Lesnik KL, Liu H (2014) Establishing a core microbiome in acetate-fed microbial fuel cells. Appl Microbiol Biotechnol 98:4187–4196. https://doi.org/10.1007/s00253-013-5502-9
Regueiro L, Carballa M, Lema JM (2014) Outlining microbial community dynamics during temperature drop and subsequent recovery period in anaerobic co-digestion systems. J Biotechnol 192:179–186. https://doi.org/10.1016/j.jbiotec.2014.10.007
Dahle H, Birkeland N-K (2006) Thermovirga lienii gen. Nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int J Syst Evol Microbiol 56:1539–1545. https://doi.org/10.1099/ijs.0.63894-0
Honda T, Fujita T, Tonouchi A (2013) Aminivibrio pyruvatiphilus gen. Nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int J Syst Evol Microbiol 63:3679–3686. https://doi.org/10.1099/ijs.0.052225-0
Meng X, Yuan X, Ren J, Wang X, Zhu W, Cui Z (2017) Methane production and characteristics of the microbial community in a two-stage fixed-bed anaerobic reactor using molasses. Bioresour Technol 241:1050–1059. https://doi.org/10.1016/j.biortech.2017.05.181
Ito T, Yoshiguchi K, Ariesyady HD, Okabe S (2011) Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J 5:1844–1856. https://doi.org/10.1038/ismej.2011.59
Dassa B, Borovok I, Lamed R, Henrissat B, Coutinho P, Hemme CL, Yue H, Zhou J, Bayer EA (2012) Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics 13:210. https://doi.org/10.1186/1471-2164-13-210
Xu S, Han R, Zhang Y, He C, Liu H (2018) Differentiated stimulating effects of activated carbon on methanogenic degradation of acetate, propionate and butyrate. Waste Manag 76:394–403. https://doi.org/10.1016/j.wasman.2018.03.037
Lee SH, Park JH, Kang HJ, Lee YH, Lee TJ, Park HD (2013) Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresour Technol 145:25–32. https://doi.org/10.1016/j.biortech.2013.02.070
Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3:700–714. https://doi.org/10.1038/ismej.2009.2
Wang HZ, Gou M, Yi Y, Xia ZY, Tang YQ (2018) Identification of novel potential acetate-oxidizing bacteria in an acetate-fed methanogenic chemostat based on DNA stable isotope probing. J Gen Appl Microbiol 64:221–231. https://doi.org/10.2323/jgam.2017.12.006
Hao L, Fan L, Mazéas L, Quéméner DL, Madigou C, Guenne A, Shao L, Bouchez T, He P (2015) Stable isotope probing of acetate fed anaerobic batch incubations shows a partial resistance of acetoclastic methanogenesis catalyzed by Methanosarcina to sudden increase of ammonia level. Water Res 69:90–99. https://doi.org/10.1016/j.watres.2014.11.010
Neubeck A, Sjöberg S, Price A, Callac N, Schnürer A (2016) Effect of nickel levels on hydrogen partial pressure and methane production in methanogens. PLoS One 11:e0168357. https://doi.org/10.1371/journal.pone.0168357
Zinder SH (1993) Physiological ecology of methanogens. Springer, US
Worm P, Koehorst JJ, Visser M, Sedanonúñez VT, Schaap PJ, Plugge CM, Sousa DZ, Stams AJ (2014) A genomic view on syntrophic versus non-syntrophic lifestyle in anaerobic fatty acid degrading communities. Biochim Biophys Acta 1837:2004–2016. https://doi.org/10.1016/j.bbabio.2014.06.005
Funding
This study was funded by the Ministry of Science and Technology of China (No. 2016YFE0127700) and by the National Natural Science Foundation of China (No. 51678378; No. 31200068).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 2147 kb)
Rights and permissions
About this article
Cite this article
Yi, Y., Wang, H., Chen, Y. et al. Identification of Novel Butyrate- and Acetate-Oxidizing Bacteria in Butyrate-Fed Mesophilic Anaerobic Chemostats by DNA-Based Stable Isotope Probing. Microb Ecol 79, 285–298 (2020). https://doi.org/10.1007/s00248-019-01400-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01400-z