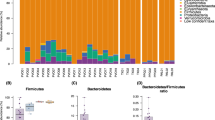
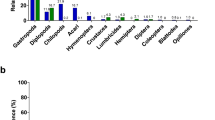
Abstract
Gut microbial communities of animals are influenced by diet and seasonal weather changes. Since foraging strategies of wild animals are affected by phenological changes, gut microbial communities would differ among seasons. However, interactions of plant-animal-microbiota with seasonal changes have not been well characterized. Here, we surveyed gut microbial diversity of Siberian flying squirrels (Pteromys volans orii) from a natural forest in Hokkaido during spring and summer of 2013 and 2014. Additionally, we compared microbial diversity to temperature changes and normalized difference vegetation index (NDVI). Changes in both seasonal temperature and phenology were significantly associated with alterations in gut microbiota. There were two clusters of OTUs, below and above 20 °C that were significantly correlated with low and high temperatures, respectively. Low-temperature cluster OTUs belonged to various phyla, whereas the high-temperature cluster was only constituted by Firmicutes. In conclusion, gut microbiota of Siberian flying squirrels varied with environmental changes on an ecological scale.



Similar content being viewed by others
References
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651. https://doi.org/10.1126/science.1155725
Kostic AD, Howitt MR, Garrett WS (2013) Exploring host–microbiota interactions in animal models and humans. Genes Dev 27:701–718. https://doi.org/10.1101/gad.212522.112
Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. https://doi.org/10.1126/scitranslmed.3000322
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. https://doi.org/10.1038/nature12820
Moschen AR, Wieser V, Tilg H (2012) Dietary factors: major regulators of the gut’s microbiota. Gut Liver 6:411–416. https://doi.org/10.5009/gnl.2012.6.4.411
Maurice CF, Knowles SC, Ladau J, Pollard KS, Fenton A, Pedersen AB, Turnbaugh PJ (2015) Marked seasonal variation in the wild mouse gut microbiota. ISME J 9:2423–2434. https://doi.org/10.1038/ismej.2015.53
Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL (2017) Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357:802–806. https://doi.org/10.1126/science.aan4834
Kohl KD, Yahn J (2016) Effects of environmental temperature on the gut microbial communities of tadpoles. Environ Microbiol 18:1561–1565. https://doi.org/10.1111/1462-2920.13255
Kikuchi Y, Tada A, Musolin DL, Hari N, Hosokawa T, Fujisaki K, Fukatsu T (2016) Collapse of insect gut symbiosis under simulated climate change. MBio 7. https://doi.org/10.1128/mBio.01578-16
Shoemaker KM, Moisander PH (2017) Seasonal variation in the copepod gut microbiome in the subtropical North Atlantic Ocean. Environ Microbiol 19:3087–3097. https://doi.org/10.1111/1462-2920.13780
Sun B, Wang X, Bernstein S, Huffman MA, Xia DP, Gu Z, Chen R, Sheeran LK, Wagner RS, Li J (2016) Marked variation between winter and spring gut microbiota in free-ranging Tibetan macaques (Macaca thibetana). Sci Rep 6:26035. https://doi.org/10.1038/srep26035
Stone AI (2007) Responses of squirrel monkeys to seasonal changes in food availability in an eastern Amazonian forest. Am J Primatol 69:142–157. https://doi.org/10.1002/ajp.20335
Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Wilson BA, Nelson KE, White BA, Garber PA (2015) The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol 69:434–443. https://doi.org/10.1007/s00248-014-0554-7
Amato KR, Ulanov A, Ju KS, Garber PA (2017) Metabolomic data suggest regulation of black howler monkey (Alouatta pigra) diet composition at the molecular level. Am J Primatol 79:1–10. https://doi.org/10.1002/ajp.22616
Siles JA, Margesin R (2017) Seasonal soil microbial responses are limited to changes in functionality at two Alpine forest sites differing in altitude and vegetation. Sci Rep 7:2204. https://doi.org/10.1038/s41598-017-02363-2
Solden LM, Hoyt DW, Collins WB, Plank JE, Daly RA, Hildebrand E, Beavers TJ, Wolfe R, Nicora CD, Purvine SO, Carstensen M, Lipton MS, Spalinger DE, Firkins JL, Wolfe BA, Wrighton KC (2017) New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J 11:691–703. https://doi.org/10.1038/ismej.2016.150
Stevenson TJ, Duddleston KN, Buck CL (2014) Effects of season and host physiological state on the diversity, density, and activity of the arctic ground squirrel cecal microbiota. Appl Environ Microbiol 80:5611–5622. https://doi.org/10.1128/AEM.01537-14
Kuo C-C, Lee L-L (2003) Food availability and food habits of Indian giant flying squirrels (Petaurista Philippensis) in Taiwan. J Mammal 84:1330–1340. https://doi.org/10.1644/bos-039
Okitsu S (2003) Forest vegetation of northern Japan and the southern Kurils. In: Kolbek J, Šrůtek M, Box EO (eds) Forest vegetation of Northeast Asia. Springer Netherlands, Dordrecht, pp 231–261
Nagai S, Nakai T, Saitoh TM, Busey RC, Kobayashi H, Suzuki R, Muraoka H, Kim Y (2013) Seasonal changes in camera-based indices from an open canopy black spruce forest in Alaska, and comparison with indices from a closed canopy evergreen coniferous forest in Japan. Polar Sci 7:125–135. https://doi.org/10.1016/j.polar.2012.12.001
Olenichenko NA, Zagoskina NV, Astakhova NV, Trunova TI, Kuznetsov YV (2008) Primary and secondary metabolism of winter wheat under cold hardening and treatment with antioxidants. Appl Biochem Microbiol 44:535–540. https://doi.org/10.1134/s0003683808050141
Evans AL, Singh NJ, Friebe A, Arnemo JM, Laske TG, Frobert O, Swenson JE, Blanc S (2016) Drivers of hibernation in the brown bear. Front Zool 13:7. https://doi.org/10.1186/s12983-016-0140-6
Chevalier C, Stojanovic O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, Rigo D, Fabbiano S, Stevanovic A, Hagemann S, Montet X, Seimbille Y, Zamboni N, Hapfelmeier S, Trajkovski M (2015) Gut microbiota orchestrates energy homeostasis during cold. Cell 163:1360–1374. https://doi.org/10.1016/j.cell.2015.11.004
Zietak M, Kovatcheva-Datchary P, Markiewicz LH, Stahlman M, Kozak LP, Backhed F (2016) Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab 23:1216–1223. https://doi.org/10.1016/j.cmet.2016.05.001
Derrien M, Vaughan EE, Plugge CM, de Vos WM (2004) Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. https://doi.org/10.1099/ijs.0.02873-0
Japan Meteorological Agency. http://www.data.jma.go.jp/obd/stats/etrn/view/nml_amd_10d.php?prec_no=12&block_no=0021&year=&month=&day=&view=a2
Kadoya N, Iguchi K, Matsui M, Okahira T, Kato A, Oshida T, Hayashi Y (2010) A preliminary survey on nest cavity use by Siberian flying squirrels, Pteromys volans orii, in forests of Hokkaido Island, Japan. Russian Journal of Theriology 9:27–32. https://doi.org/10.15298/rusjtheriol.9.1.04
Izumi I, Matsui M, Okahira T, Hayashi Y, Oshida T (2011) Preliminary survey of habitat use by Sciurus vulgaris orientis in a natural forest of Hokkaido Island, Japan. Mamm Study 36:109–112. https://doi.org/10.3106/041.036.0202
Yamamoto H, Nitami T, Kisanuki H (1995) Stand structure of mixed-species stands ( I ) Relation of species composition and topographic factors. J Jap Forest Soc 77:47–54. https://doi.org/10.11519/jjfs1953.77.1_47
Abe H, Ishii N, Itoo T, Kaneko Y, Maeda K, Miura S, Yoneda M (2005) A guide to the mammals of Japan. Tokai University Press, Kanagawa
Hanski IK, Stevens PC, Ihalempiä P, Selonen V (2000) Home-range size, movements, and nest-site use in the Siberian flying squirrel, Pteromys Volans. J Mammal 81:798–809. https://doi.org/10.1644/1545-1542(2000)081<0798:Hrsman>2.3.Co;2
Comeau AM, Douglas GM, Langille MG (2017) Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems 2. https://doi.org/10.1128/mSystems.00127-16
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:10. https://doi.org/10.14806/ej.17.1.200
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: a fast and accurate illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. https://doi.org/10.1093/bioinformatics/btt593
Rognes T, Flouri T, Nichols B, Quince C, Mahe F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R (2011) Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics Chapter 10: Unit 10 17. https://doi.org/10.1002/0471250953.bi1007s36
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Kopylova E, Noe L, Touzet H (2012) SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. https://doi.org/10.1093/bioinformatics/bts611
Mercier C, Boyer F, Bonin A, Coissac E (2013) SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. Laboratoire d’Écologie Alpine, France
Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW (2014) CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11. https://doi.org/10.1186/2049-2618-2-11
Kembel SW, Wu M, Eisen JA, Green JL (2012) Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput Biol 8:e1002743. https://doi.org/10.1371/journal.pcbi.1002743
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015) Vegan: community ecology package. R package version 2.3-1. Oulu, Finland
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Fox J, Weisberg S (2011) An {R} companion to applied regression. vol Second. Sage, Thousand Oaks
Pettorelli N, Vik JO, Mysterud A, Gaillard JM, Tucker CJ, Stenseth NC (2005) Using the satellite-derived NDVI to assess ecological responses to environmental change. Trends Ecol Evol 20:503–510. https://doi.org/10.1016/j.tree.2005.05.011
Didan K (2015) MOD13Q1 MODIS/Terra vegetation indices 16-day L3 global 250m SIN grid V006. NASA EOSDIS Land Processes DAAC
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Schwiertz A, Hold GL, Duncan SH, Gruhl B, Collins MD, Lawson PA, Flint HJ, Blaut M (2002) Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst Appl Microbiol 25:46–51. https://doi.org/10.1078/0723-2020-00096
Eeckhaut V, Van Immerseel F, Pasmans F, De Brandt E, Haesebrouck F, Ducatelle R, Vandamme P (2010) Anaerostipes butyraticus sp. nov., an anaerobic, butyrate-producing bacterium from Clostridium cluster XIVa isolated from broiler chicken caecal content, and emended description of the genus Anaerostipes. Int J Syst Evol Microbiol 60:1108–1112. https://doi.org/10.1099/ijs.0.015289-0
Bui TP, de Vos WM, Plugge CM (2014) Anaerostipes rhamnosivorans sp. nov., a human intestinal, butyrate-forming bacterium. Int J Syst Evol Microbiol 64:787–793. https://doi.org/10.1099/ijs.0.055061-0
Pough FH, Janis CM, Heiser JB (2013) Vertebrate life. Pearson International Edition
Bestion E, Jacob S, Zinger L, Di Gesu L, Richard M, White J, Cote J (2017) Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat Ecol Evol 1:161. https://doi.org/10.1038/s41559-017-0161
Suzuki TA (2017) Links between natural variation in the microbiome and host fitness in wild mammals. Integr Comp Biol 57:756–769. https://doi.org/10.1093/icb/icx104
Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman W (2011) Bergey’s manual of systematic bacteriology: volume 3: the firmicutes. Springer Science & Business Media
Itoh T, Iino T (2013) Phylogeny and biological features of thermophiles. In: Satyanarayana T, Littlechild J, Kawarabayasi Y (eds) Thermophilic microbes in environmental and industrial biotechnology: biotechnology of thermophiles. Springer Netherlands, Dordrecht, pp 249–270
Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-i, Ludwig W, Whitman WB (2012) Bergey’s manual of systematic bacteriology: volume 5: the actinobacteria. Springer-Verlag, New York
Cho GS, Ritzmann F, Eckstein M, Huch M, Briviba K, Behsnilian D, Neve H, Franz CM (2016) Quantification of Slackia and Eggerthella spp. in human feces and adhesion of representatives strains to Caco-2 cells. Front Microbiol 7:658. https://doi.org/10.3389/fmicb.2016.00658
Clavel T, Henderson G, Alpert CA, Philippe C, Rigottier-Gois L, Dore J, Blaut M (2005) Intestinal bacterial communities that produce active estrogen-like compounds enterodiol and enterolactone in humans. Appl Environ Microbiol 71:6077–6085. https://doi.org/10.1128/AEM.71.10.6077-6085.2005
Clavel T, Henderson G, Engst W, Dore J, Blaut M (2006) Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside. FEMS Microbiol Ecol 55:471–478. https://doi.org/10.1111/j.1574-6941.2005.00057.x
Jin J-S, Zhao Y-F, Nakamura N, Akao T, Kakiuchi N, Min B-S, Hattori M (2007) Enantioselective dehydroxylation of enterodiol and enterolactone precursors by human intestinal bacteria. Biol Pharm Bull 30:2113–2119. https://doi.org/10.1248/bpb.30.2113
Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, Ley RE (2013) The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2:e01102. https://doi.org/10.7554/eLife.01102
Zeng B, Han S, Wang P, Wen B, Jian W, Guo W, Yu Z, Du D, Fu X, Kong F, Yang M, Si X, Zhao J, Li Y (2015) The bacterial communities associated with fecal types and body weight of rex rabbits. Sci Rep 5:9342. https://doi.org/10.1038/srep09342
Acknowledgements
We thank Hao-Ting Chang for guidance regarding analysis of normalized difference vegetation index, A. Sanyoshi and K. Iguchi of the University of Tokyo Hokkaido Forest for their cooperation in the field, and John Wang for helpful comments for this manuscript.
Funding
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 103-2311-B-002-001 and MOST 106-2633-B-006-004).
Author information
Authors and Affiliations
Contributions
P-YL, A-CC, and H-TY conceived the study design; PY-L, HW-C, TO, and H-TY collected sample; P-YL, A-CC, and S-WH conducted experiments; P-YL and A-CC conducted bioinformatics analyses; P-YL, A-CC, S-WH, and H-TY wrote the first draft. All authors contributed to data interpretation and preparation of the final manuscript. All authors reviewed and approved the final manuscript.
Corresponding author
Electronic Supplementary Material
ESM 1
(PDF 541 kb)
Rights and permissions
About this article
Cite this article
Liu, PY., Cheng, AC., Huang, SW. et al. Variations in Gut Microbiota of Siberian Flying Squirrels Correspond to Seasonal Phenological Changes in Their Hokkaido Subarctic Forest Ecosystem. Microb Ecol 78, 223–231 (2019). https://doi.org/10.1007/s00248-018-1278-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1278-x