Abstract
Ephemeral aquatic habitats and their associated microbial communities (microbiomes) play important roles in the growth and development of numerous aquatic insects, including mosquitoes (Diptera). Biological control agents, such as Bacillus thuringiensis israelensis (Bti) or insect growth regulators (e.g., methoprene), are commonly used to control mosquitoes in these habitats. However, it is unknown how commonly used control compounds affect the mosquito internal microbiome and potentially alter their life history traits. The objectives of this study were threefold: characterize the internal microbiota of Aedes larvae (Culicidae) in ephemeral forested mosquito habitat using high-throughput amplicon based sequencing, assess how mosquito control treatments affect the internal microbial communities of larval mosquitoes, and determine if changes to the microbiome resulted from direct or indirect treatment effects. The larval microbiome varied in community composition and diversity with development stage and treatment, suggesting potential effects of control compounds on insect microbial ecology. While microbial community differences due to Bti treatment were a result of indirect effects on larval development, methoprene had significant impacts on bacterial and algal taxa that could not be explained by indirect treatment effects. These results provide new information on the interactions between pesticide treatments and insect microbial communities.




Similar content being viewed by others
References
Banning NC, Gleeson DB, Grigg AH, Grant CD, Andersen GL, Brodie EL, Murphy DV (2011) Soil microbial community successional patterns during Forest ecosystem restoration. Appl Environ Microbiol 77:6158–6164. https://doi.org/10.1128/AEM.00764-11
Hättenschwiler S, Coq S, Barantal S, Handa IT (2011) Leaf traits and decomposition in tropical rainforests: revisiting some commonly held views and towards a new hypothesis. New Phytol. 189:950–965. https://doi.org/10.1111/j.1469-8137.2010.03483.x
Zak DR, Holmes WE, White DC, Peacock AD, Tilman D (2003) Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology 84:2042–2050. https://doi.org/10.1890/02-0433
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92. https://doi.org/10.1146/annurev.ento.49.061802.123416
Coon KL, Vogel KJ, Brown MR, Strand MR (2014) Mosquitoes rely on their gut microbiota for development. Mol Ecol 23:2727–2739. https://doi.org/10.1111/mec.12771
Merritt RW, Dadd RH, Walker ED (1992) Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Annu Rev Entomol 37:349–376. https://doi.org/10.1146/annurev.en.37.010192.002025
Ponnusamy L, Xu N, Nojima S, Wesson DM, Schal C, Apperson CS (2008) Identification of bacteria and bacteria-associated chemical cues that mediate oviposition site preferences by Aedes aegypti. Proc Natl Acad Sci U S A 105:9262–9267. https://doi.org/10.1073/pnas.0802505105
Wallace JR, Merritt RW (2004) Diel feeding periodicity of larval Anopheline mosquitoes on microorganisms and Microinvertebrates: a spatial and temporal comparison of Anopheles quadrimaculatus (Diptera: Culicidae) diets in a Michigan pond. J Med Entomol 41:853–860. https://doi.org/10.1603/0022-2585-41.5.853
Kaufman MG, Walker ED, Smith TW, Merritt RW, Klug MJ (1999) Effects of larval mosquitoes (Aedes triseriatus) and Stemflow on microbial community dynamics in container habitats. Appl Environ Microbiol 65:2661–2673
Walker ED, Kaufman MG, Merritt RW (2010) An acute trophic cascade among microorganisms in the tree hole ecosystem following removal of omnivorous mosquito larvae. Community Ecol : CE 11:171–178. https://doi.org/10.1556/ComEc.11.2010.2.5
Pernthaler J (2005) Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546
Muturi EJ, Ramirez JL, Rooney AP, Kim C-H (2017) Comparative analysis of gut microbiota of mosquito communities in Central Illinois. PLoS Negl Trop Dis 11:e0005377. https://doi.org/10.1371/journal.pntd.0005377
Charan SS, Pawar KD, Severson DW, Patole MS, Shouche YS (2013) Comparative analysis of midgut bacterial communities of Aedes aegypti mosquito strains varying in vector competence to dengue virus. Parasitol Res 112:2627–2637. https://doi.org/10.1007/s00436-013-3428-x
David MR, Santos LM, Vicente AC, Maciel-de-Freitas R (2016) Effects of environment, dietary regime and ageing on the dengue vector microbiota: evidence of a core microbiota throughout Aedes aegypti lifespan. Mem Inst Oswaldo Cruz 111:577–587. https://doi.org/10.1590/0074-02760160238
Kim CH, Lampman RL, Muturi EJ (2015) Bacterial communities and midgut microbiota associated with mosquito populations from waste tires in East-Central Illinois. J Med Entomol 52:63–75. https://doi.org/10.1093/jme/tju011
Walker ED, Olds EJ, Merritt RW (1988) Gut content analysis of mosquito larvae (Diptera: Culicidae) using Dapi stain and epifluorescence microscopy. J Med Entomol 25:551–554. https://doi.org/10.1093/jmedent/25.6.551
Dong Y, Manfredini F, Dimopoulos G (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5:e1000423. https://doi.org/10.1371/journal.ppat.1000423
Cirimotich CM, Ramirez JL, Dimopoulos G (2011) Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 10:307–310. https://doi.org/10.1016/j.chom.2011.09.006
Bian G, Zhou G, Lu P, Xi Z (2013) Replacing a native Wolbachia with a novel strain results in an increase in endosymbiont load and resistance to dengue virus in a mosquito vector. PLoS Negl Trop Dis 7:e2250. https://doi.org/10.1371/journal.pntd.0002250
Xi Z, Gavotte L, Xie Y, Dobson SL (2008) Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genomics 9(1):1. https://doi.org/10.1186/1471-2164-9-1
Wipfli MS, Merritt RW (1994) Disturbance to a stream food web by a bacterial larvicide specific to black flies: feeding responses of predatory macroinvertebrates. Freshwat Biol 32:91–103. https://doi.org/10.1111/j.1365-2427.1994.tb00869.x
Lacey LA (2007) Bacillus thuringiensis serovariety israelensis and bacillus sphaericus for mosquito control. J Am Mosq Control Assoc 23:133–163. https://doi.org/10.2987/8756-971X(2007)23[133:BTSIAB]2.0.CO;2
Pruszynski CA, Hribar LJ, Mickle R, Leal AL (2017) A large scale Biorational approach using Bacillus thuringiensis israeliensis (strain AM65-52) for managing Aedes aegypti populations to prevent dengue, chikungunya and Zika transmission. PLoS One 12:e0170079. https://doi.org/10.1371/journal.pone.0170079
Butler M, Lebrun RA, Ginsberg HS, Gettman AD (2006) Efficacy of methoprene for mosquito control in storm water catch basins. J Am Mosq Control Assoc 22:333–338. https://doi.org/10.2987/8756-971x(2006)22[333:eomfmc]2.0.co;2
Chilcott CN, Knowles BH, Ellar DJ, Drobniewski FA (1990) Mechanism of action of Bacillus thuringiensis israelensis parasporal body. In: de Barjac H, Sutherland DJ (eds) Bacterial Control of Mosquitoes & Black Flies: Biochemistry, Genetics & Applications of Bacillus thuringiensis israelensis and Bacillus sphaericus. Springer Netherlands, Dordrecht, pp 45–65
Loschiavo SR (1976) Effects of the synthetic insect growth regulators methoprene and hydroprene on survival, development or reproduction of six species of stored-products insects 12. J Econ Entomol 69:395–399. https://doi.org/10.1093/jee/69.3.395
Lacey LA, Merritt RW (2003) The safety of bacterial microbial agents used for black fly and mosquito control in aquatic environments. In: Hokkanen HMT, Hajek AE (eds) Environmental impacts of microbial insecticides: need and methods for risk assessment. Springer Netherlands, Dordrecht, pp 151–168
Duguma D, Hall MW, Rugman-Jones P, Stouthamer R, Neufeld JD, Walton WE (2015) Microbial communities and nutrient dynamics in experimental microcosms are altered after the application of a high dose of Bti. J Appl Ecol 52:763–773. https://doi.org/10.1111/1365-2664.12422
Muturi EJ, Orindi BO, Kim C-H (2013) Effect of leaf type and pesticide exposure on abundance of bacterial taxa in mosquito larval habitats. PLoS One 8:e71812. https://doi.org/10.1371/journal.pone.0071812
Muturi EJ, Donthu RK, Fields CJ, Moise IK, Kim C-H (2017) Effect of pesticides on microbial communities in container aquatic habitats. Sci Rep 7:44565. https://doi.org/10.1038/srep44565
Darsie RF, Ward RA (2005) Identification and geographical distribution of the mosquitos of North America, North of Mexico
Schindelin J, Rueden CT, Hiner MC, Eliceiri KW (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. https://doi.org/10.1002/mrd.22489
Ridley EV, Wong ACN, Westmiller S, Douglas AE (2012) Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One 7:e36765. https://doi.org/10.1371/journal.pone.0036765
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O'Connor M, Harnedy N, O'Connor K, Henry C, O'Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O'Toole PW (2010) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 108
Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R (2011) Moving pictures of the human microbiome. Genome Biol 12:R50. https://doi.org/10.1186/gb-2011-12-5-r50
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Pechal JL, Benbow ME (2016) Microbial ecology of the salmon necrobiome: evidence salmon carrion decomposition influences aquatic and terrestrial insect microbiomes. Environ Microbiol 18:1511–1522. https://doi.org/10.1111/1462-2920.13187
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. https://doi.org/10.1038/nmeth.2276
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. https://doi.org/10.1101/gr.112730.110
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/aem.00062-07
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/aem.03006-05
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J 6:610–618. https://doi.org/10.1038/ismej.2011.139
Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE (2012) Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. Isme J 6:94–103. https://doi.org/10.1038/ismej.2011.82
Lehmann K, Singer A, Bowes MJ, Ings NL, Field D, Bell T (2015) 16S rRNA assessment of the influence of shading on early-successional biofilms in experimental streams. FEMS Microbiol. Ecol. 91:fiv129. https://doi.org/10.1093/femsec/fiv129
Fox J, Weisberg S (2011) An R companion to applied regression. Sage, Thousand Oaks
team Rcd RA Language and Environment for Statistical Computing (2014), R Foundation for Statistical Computing, Vienna, Austria, R foundation for statistical computing. ISBN 3-900051-07-0
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. https://doi.org/10.1111/j.1442-9993.2001.01070.pp.x
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Liaw A, Wiener M (2002) Classification and regression by randomForest. R news 2:18–22
McDaniel IN, Horsfall WR (1963) Bionomics of Aedes stimulans (Diptera: Culicidae) I. Effect of moisture on the distribution of eggs. Am Midl Nat 70:479–489. https://doi.org/10.2307/2423072
Podrabsky JE, Hrbek T, Hand SC (1997) Physical and chemical characteristics of ephemeral pond habitats in the Maracaibo basin and Llanos region of Venezuela. Hydrobiologia 362:67–77. https://doi.org/10.1023/a:1003168704178
Carrino-Kyker SR, Swanson AK (2008) Temporal and spatial patterns of eukaryotic and bacterial communities found in vernal pools. Appl Environ Microbiol 74:2554–2557. https://doi.org/10.1128/AEM.01482-07
Duguma D, Hall MW, Rugman-Jones P, Stouthamer R, Terenius O, Neufeld JD, Walton WE (2015) Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol 15:140. https://doi.org/10.1186/s12866-015-0475-8
Wang Y, Gilbreath TM, Kukutla P, Yan G, Xu J (2011) Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6:e24767. https://doi.org/10.1371/journal.pone.0024767
Brabant PJ, Dobson SL (2013) Methoprene effects on survival and reproductive performance of adult female and male Aedes aegypti. J Am Mosq Control Assoc 29:369–375. https://doi.org/10.2987/13-6365.1
Bai H, Gelman DB, Palli SR (2010) Mode of action of methoprene in affecting female reproduction in the African malaria mosquito, Anopheles gambiae. Pest Manag Sci 66:936–943. https://doi.org/10.1002/ps.1962
Tetreau G, Stalinski R, Kersusan D, Veyrenc S, David J-P, Reynaud S, Després L (2012) Decreased toxicity of Bacillus thuringiensis subsp. israelensis to mosquito larvae after contact with leaf litter. Appl Environ Microbiol 78:5189–5195
Duguma D, Rugman-Jones P, Kaufman MG, Hall MW, Neufeld JD, Stouthamer R, Walton WE (2013) Bacterial communities associated with Culex Mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS One 8:e72522. https://doi.org/10.1371/journal.pone.0072522
Pennington MJ, Prager SM, Walton WE, Trumble JT (2016) Culex quinquefasciatus larval microbiomes vary with instar and exposure to common wastewater contaminants6: 21969. doi: https://doi.org/10.1038/srep21969
Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, Shahbazkia HR, Awono-Ambene PH, Levashina EA, Christen R, Morlais I (2012) Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog 8:e1002742. https://doi.org/10.1371/journal.ppat.1002742
Minard G, Tran FH, Raharimalala FN, Hellard E, Ravelonandro P, Mavingui P, Valiente MC (2013) Prevalence, genomic and metabolic profiles of Acinetobacter and Asaia associated with field-caught Aedes albopictus from Madagascar. FEMS Microbiol Ecol 83:63–73. https://doi.org/10.1111/j.1574-6941.2012.01455.x
Acknowledgements
We would like to thank Matthew Silva, Greg Vaccarino, Frank Herr, Ryan Walker, and Jon Rutt for assistance in the field; Courtney Weatherbee for assistance with DNA extraction; and Chris Hardy, Brent Horton, and Sepi Yalda, who served on JRs thesis committee. Funding support for this project was provided by Hunterdon County Vector Control Program (HCVCP) black fly grant no. 6032305751, Commonwealth of Pennsylvania University Biologists (CPUB) Student Research Grant, Neimeyer-Hodgson Student Research Grant, Noonan Endowment award, and William Yurkiewicz Fellowship. We also thank the College of Agriculture and Natural Resources (Department of Entomology) and the College of Osteopathic Medicine (Department of Osteopathic Medical Specialties) for funding this work (MEB).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Table S1
Samples for gene amplicon sequencing by instar and date. (DOCX 12 kb)
Table S2
Samples for gene amplicon sequencing by treatment and instar (DOCX 12 kb)
Table S3
Most important predictors of treatment group between methoprene and control samples at greater than one day post. (DOCX 13 kb)
Figure S1
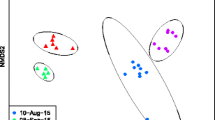
Principle coordinate analysis by sampling date. (GIF 101 kb)
Figure S2
Locations of sample divots within Teetertown preserve. (GIF 1079 kb)
Rights and permissions
About this article
Cite this article
Receveur, J.P., Pechal, J.L., Benbow, M.E. et al. Changes in Larval Mosquito Microbiota Reveal Non-target Effects of Insecticide Treatments in Hurricane-Created Habitats. Microb Ecol 76, 719–728 (2018). https://doi.org/10.1007/s00248-018-1175-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1175-3