Abstract
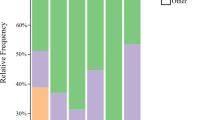
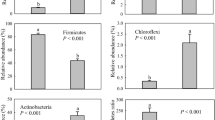
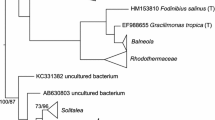
Blood feeding red poultry mites (RPM) serve as vectors of pathogenic bacteria and viruses among vertebrate hosts including wild birds, poultry hens, mammals, and humans. The microbiome of RPM has not yet been studied by high-throughput sequencing. RPM eggs, larvae, and engorged adult/nymph samples obtained in four poultry houses in Czechia were used for microbiome analyses by Illumina amplicon sequencing of the 16S ribosomal RNA (rRNA) gene V4 region. A laboratory RPM population was used as positive control for transcriptome analysis by pyrosequencing with identification of sequences originating from bacteria. The samples of engorged adult/nymph stages had 100-fold more copies of 16S rRNA gene copies than the samples of eggs and larvae. The microbiome composition showed differences among the four poultry houses and among observed developmental stadia. In the adults’ microbiome 10 OTUs comprised 90 to 99% of all sequences. Bartonella-like bacteria covered between 30 and 70% of sequences in RPM microbiome and 25% bacterial sequences in transcriptome. The phylogenetic analyses of 16S rRNA gene sequences revealed two distinct groups of Bartonella-like bacteria forming sister groups: (i) symbionts of ants; (ii) Bartonella genus. Cardinium, Wolbachia, and Rickettsiella sp. were found in the microbiomes of all tested stadia, while Spiroplasma eriocheiris and Wolbachia were identified in the laboratory RPM transcriptome. The microbiomes from eggs, larvae, and engorged adults/nymphs differed. Bartonella-like symbionts were found in all stadia and sampling sites. Bartonella-like bacteria was the most diversified group within the RPM microbiome. The presence of identified putative pathogenic bacteria is relevant with respect to human and animal health issues while the identification of symbiontic bacteria can lead to new control methods targeting them to destabilize the arthropod host.




Similar content being viewed by others
References
Pritchard J, Kuster T, Sparagano O, Tomley F (2015) Understanding the biology and control of the poultry red mite Dermanyssus gallinae: a review. Avian Pathol 44:143–153. doi:10.1080/03079457.2015.1030589
Lucky AW, Sayers CP, Argus JD, Lucky A (2001) Avian mite bites acquired from a new source—pet gerbils: report of 2 cases and review of the literature. Arch Dermatol 137:167–170. doi:10.1001/pubs.Arch Dermatol.-ISSN-0003-987x-137-2-dob00013
Cafiero MA, Camarda A, Circella E, Santagada G, Schino G, Lomuto M (2008) Pseudoscabies caused by Dermanyssus gallinae in Italian city dwellers: a new setting for an old dermatitis. J Eur Acad Dermatol Venereol 22:1382–1383. doi:10.1111/j.1468-3083.2008.02645.x
Sparagano OAE, George DR, Harrington DWJ, Giangaspero A (2014) Significance and control of the poultry red mite, Dermanyssus gallinae. Annu Rev Entomol 59:447–466. doi:10.1146/annurev-ento-011613-162101
Moro CV, De Luna CJ, Tod A, Guy JH, Sparagano OAE, Zenner L (2009) The poultry red mite (Dermanyssus gallinae): a potential vector of pathogenic agents. Exp Appl Acarol 48:93–104. doi:10.1007/s10493-009-9248-0
Moro CV, Fravalo P, Amelot M, Chauve C, Zenner L, Salvat G (2007) Colonization and organ invasion in chicks experimentally infected with Dermanyssus gallinae contaminated by Salmonella enteritidis. Avian Pathol 36:307–311. doi:10.1080/03079450701460484
Moro CV, Thioulouse J, Chauve C, Normand P, Zenner L (2009) Bacterial taxa associated with the hematophagous mite Dermanyssus gallinae detected by 16S rRNA PCR amplification and TTGE fingerprinting. Res Microbiol 160:63–70. doi:10.1016/j.resmic.2008.10.006
Rasmussen M (2016) Aerococcus: an increasingly acknowledged human pathogen. Clin Microbiol Infect 22:22–27. doi:10.1016/j.cmi.2015.09.026
De Luna CJ, Arkle S, Harrington D, George DR, Guy JH, Sparagano OAE (2008) The poultry red mite Dermanyssus gallinae as a potential carrier of vector-borne diseases. Ann N Y Acad Sci 1149:255–258. doi:10.1196/annals.1428.085
Huong CTT, Murano T, Uno Y, Usui T, Yamaguchi T (2014) Molecular detection of avian pathogens in poultry red mite (Dermanyssus gallinae) collected in chicken farms. J Vet Med Sci 76:1583–1587. doi:10.1292/jvms.14-0253
Circella E, Pugliese N, Todisco G, Cafiero MA, Sparagano OAE, Camarda A (2011) Chlamydia psittaci infection in canaries heavily infested by Dermanyssus gallinae. Exp Appl Acarol 55:329–338. doi:10.1007/s10493-011-9478-9
De Luna CJ, Moro CV, Guy JH, Zenner L, Sparagano OAE (2009) Endosymbiotic bacteria living inside the poultry red mite (Dermanyssus gallinae). Exp Appl Acarol 48:105–113. doi:10.1007/s10493-008-9230-2
Hodkinson BP, Grice EA (2015) Next-generation sequencing: a review of technologies and tools for wound microbiome research. Adv Wound Care (New Rochelle) 4:50–58. doi:10.1089/wound.2014.0542
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145. doi:10.1038/nbt1486
Mul M, van Niekerk T, Chirico J, Maurer V, Kilpinen O, Sparagano O, Thind B, Zoons J, Moore D, Bell B, Gjevre A-G, Chauve C (2009) Control methods for Dermanyssus gallinae in systems for laying hens: results of an international seminar. World Poultry Sci J 65:589–599. doi:10.1017/S0043933909000403
Schicht S, Qi W, Poveda L, Strube C (2014) Whole transcriptome analysis of the poultry red mite Dermanyssus gallinae (de Geer, 1778). Parasitology 141:336–346. doi:10.1017/S0031182013001467
Hubert J, Erban T, Kamler M, Kopecky J, Nesvorna M, Hejdankova S, Titera D, Tyl J, Zurek L (2015) Bacteria detected in the honeybee parasitic mite Varroa destructor collected from beehive winter debris. J Appl Microbiol 119:640–654. doi:10.1111/jam.12899
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, pp. 115–175
Kopecky J, Perotti MA, Nesvorna M, Erban T, Hubert J (2013) Cardinium endosymbionts are widespread in synanthropic mite species (Acari: Astigmata). J Invertebr Pathol 112:20–23. doi:10.1016/j.jip.2012.11.001
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736. doi:10.1128/AEM.71.12.7724-7736.2005
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741. doi:10.1128/AEM.00556-06
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi:10.1038/nmeth.2604
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi:10.1128/AEM.00062-07
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Pruesse E, Peplies J, Glockner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi:10.1093/bioinformatics/bts252
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi:10.1080/10635150390235520
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772–772. doi:10.1038/nmeth.2109
Rodrigue N, Lartillot N (2014) Site-heterogeneous mutation-selection models within the PhyloBayes-MPI package. Bioinformatics 30:1020–1021. doi:10.1093/bioinformatics/btt729
Jow H, Hudelot C, Rattray M, Higgs PG (2002) Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Mol Biol Evol 19:1591–1601. doi:10.1093/oxfordjournals.molbev.a004221
Lartillot N, Lepage T, Blanquart S (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. doi:10.1093/bioinformatics/btp368
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi:10.1093/sysbio/syq010
Rambaut A (2007) FigTree, a graphical viewer of phylogenetic trees. Molecular evolution, phylogenetics and epidemiology: research, software and publications of Andrew Rambaut and members of his research group. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 27 July 2015
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi:10.1038/ismej.2012.8
Aizenberg-Gershtein Y, Izhaki I, Santhanam R, Kumar P, Baldwin IT, Halpern M (2015) Pyridine-type alkaloid composition affects bacterial community composition of floral nectar. Sci Rep 5:11536. doi:10.1038/srep11536
Earley ZM, Akhtar S, Green SJ, Naqib A, Khan O, Cannon AR, Hammer AM, Morris NL, Li X, Eberhardt JM, Gamelli RL, Kennedy RH, Choudhry MA (2015) Burn injury alters the intestinal microbiome and increases gut permeability and bacterial translocation. PLoS One 10:e0129996. doi:10.1371/journal.pone.0129996
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi:10.1093/nar/gkt1244
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/AEM.01541-09
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi:10.1128/AEM.01043-13
Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:4 http://palaeo-electronica.org/2001_1/past/issue1_01.htm. Accessed 6 August 2016
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12:385. doi:10.1186/1471-2105-12-385
Erban T, Ledvinka O, Nesvorna M, Hubert J (2017) Experimental manipulation shows a greater influence of population than dietary perturbation on the microbiome of Tyrophagus putrescentiae. Appl Environ Microbiol 83. doi:10.1128/AEM.00128-17
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2016. vegan: Community Ecology Package. CRAN - The Comprehensive R Archive Network. http://CRAN.R-project.org/package=vegan. Accessed 6 August 2016
Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B (2016) gplots: Various R programming tools for plotting data. CRAN - The Comprehensive R Archive Network. https://CRAN.R-project.org/package=gplots. Accessed 6 August 2016
Anderson MJ, Ellingsen KE, McArdle BH (2006) Multivariate dispersion as a measure of beta diversity. Ecol Lett 9:683–693. doi:10.1111/j.1461-0248.2006.00926.x
White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:e1000352. doi:10.1371/journal.pcbi.1000352
Dorn-In S, Bassitta R, Schwaiger K, Bauer J, Holzel CS (2015) Specific amplification of bacterial DNA by optimized so-called universal bacterial primers in samples rich of plant DNA. J Microbiol Methods 113:50–56. doi:10.1016/j.mimet.2015.04.001
Hubert J, Kopecky J, Nesvorna M, Perotti MA, Erban T (2016) Detection and localization of Solitalea-like and Cardinium bacteria in three Acarus siro populations (Astigmata: Acaridae). Exp Appl Acarol 70:309–327. doi:10.1007/s10493-016-0080-z
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi:10.1093/bioinformatics/bti610
Duron O (2013) Lateral transfers of insertion sequences between Wolbachia, Cardinium and Rickettsia bacterial endosymbionts. Heredity 111:330–337. doi:10.1038/hdy.2013.56
Matsuura Y, Kikuchi Y, Meng XY, Koga R, Fukatsu T (2012) Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl Environ Microbiol 78:4149–4156. doi:10.1128/AEM.00673-12
Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE (2009) Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci U S A 106:21236–21241. doi:10.1073/pnas.0907926106
Anderson KE, Russell JA, Moreau CS, Kautz S, Sullam KE, Hu Y, Basinger U, Mott BM, Buck N, Wheeler DE (2012) Highly similar microbial communities are shared among related and trophically similar ant species. Mol Ecol 21:2282–2296. doi:10.1111/j.1365-294X.2011.05464.x
Hu Y, Lukasik P, Moreau CS, Russell JA (2014) Correlates of gut community composition across an ant species (Cephalotes varians) elucidate causes and consequences of symbiotic variability. Mol Ecol 23:1284–1300. doi:10.1111/mec.12607
Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, Zhang P, Huang Z, Berger SL, Reinberg D, Wang J, Liebig J (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329:1068–1071. doi:10.1126/science.1192428
Stoll S, Gadau J, Gross R, Feldhaar H (2007) Bacterial microbiota associated with ants of the genus Tetraponera. Biol J Linn Soc 90:399–412. doi:10.1111/j.1095-8312.2006.00730.x
Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA (2011) A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20:619–628. doi:10.1111/j.1365-294X.2010.04959.x
Kesnerova L, Moritz R, Engel P (2016) Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Microbiol 66:414–421. doi:10.1099/ijsem.0.000736
Hubert J, Kopecky J, Perotti MA, Nesvorna M, Braig HR, Sagova-Mareckova M, Macovei L, Zurek L (2012) Detection and identification of species-specific bacteria associated with synanthropic mites. Microb Ecol 63:919–928. doi:10.1007/s00248-011-9969-6
Hubert J, Nesvorna M, Kopecky J, Sagova-Mareckova M, Poltronieri P (2015) Carpoglyphus lactis (Acari: Astigmata) from various dried fruits differed in associated micro-organisms. J Appl Microbiol 118:470–484. doi:10.1111/jam.12714
Kopecky J, Nesvorna M, Hubert J (2014) Bartonella-like bacteria carried by domestic mite species. Exp Appl Acarol 64:21–32. doi:10.1007/s10493-014-9811-1
Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Comparative Sequence Program NISC, Murray PR, Turner ML, Segre JA (2012) Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22:850–859. doi:10.1101/gr.131029.111
Tsuchida T, Koga R, Fujiwara A, Fukatsu T (2014) Phenotypic effect of “Candidatus Rickettsiella viridis,” a facultative symbiont of the pea aphid (Acyrthosiphon pisum), and its interaction with a coexisting symbiont. Appl Environ Microbiol 80:525–533. doi:10.1128/AEM.03049-13
Lukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ (2013) Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett 16:214–218. doi:10.1111/ele.12031
Moquin SA, Garcia JR, Brantley SL, Takacs-Vesbach CD, Shepherd UL (2012) Bacterial diversity of bryophyte-dominant biological soil crusts and associated mites. J Arid Environ 87:110–117. doi:10.1016/j.jaridenv.2012.05.004
Moro CV, Thioulouse J, Chauve C, Zenner L (2011) Diversity, geographic distribution, and habitat-specific variations of microbiota in natural populations of the chicken mite, Dermanyssus gallinae. J Med Entomol 48:788–796. doi:10.1603/ME10113
Erban T, Klimov PB, Smrz J, Phillips TW, Nesvorna M, Kopecky J, Hubert J (2016) Populations of stored product mite Tyrophagus putrescentiae differ in their bacterial communities. Front Microbiol 7:1046. doi:10.3389/fmicb.2016.01046
Hubert J, Kopecky J, Sagova-Mareckova M, Nesvorna M, Zurek L, Erban T (2016) Assessment of bacterial communities in thirteen species of laboratory-cultured domestic mites (Acari: Acaridida). J Econ Entomol 109:1887–1896. doi:10.1093/jee/tow089
Regier Y, O’Rourke F, Kempf VA (2016) Bartonella spp.—a chance to establish one health concepts in veterinary and human medicine. Parasit Vectors 9:261. doi:10.1186/s13071-016-1546-x
Reeves WK, Dowling APG, Dasch GA (2006) Rickettsial agents from parasitic Dermanyssoidea (Acari: Mesostigmata). Exp Appl Acarol 38:181–188. doi:10.1007/s10493-006-0007-1
Klangthong K, Promsthaporn S, Leepitakrat S, Schuster AL, McCardle PW, Kosoy M, Takhampunya R (2015) The distribution and diversity of Bartonella species in rodents and their ectoparasites across Thailand. PLoS One 10:e0140856. doi:10.1371/journal.pone.0140856
Melter O, Arvand M, Votypka J, Hulinska D (2012) Bartonella quintana transmission from mite to family with high socioeconomic status. Emerg Infect Dis 18:163–165. doi:10.3201/eid1801.110186
Larson HK, Goffredi SK, Parra EL, Vargas O, Pinto-Tomas AA, McGlynn TP (2014) Distribution and dietary regulation of an associated facultative Rhizobiales-related bacterium in the omnivorous giant tropical ant, Paraponera clavata. Naturwissenschaften 101:397–406. doi:10.1007/s00114-014-1168-0
Valerio CR, Murray P, Arlian LG, Slater JE (2005) Bacterial 16S ribosomal DNA in house dust mite cultures. J Allergy Clin Immunol 116:1296–1300. doi:10.1016/j.jaci.2005.09.046
Kosoy M, Hayman DTS, Chan K-S (2012) Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol 12:894–904. doi:10.1016/j.meegid.2012.03.005
Tsai Y-L, Chang C-C, Chuang S-T, Chomel BB (2011) Bartonella species and their ectoparasites: selective host adaptation or strain selection between the vector and the mammalian host? Comp Immunol Microbiol Infect Dis 34:299–314. doi:10.1016/j.cimid.2011.04.005
Mediannikov O, Sekeyova Z, Birg M-L, Raoult D (2010) A novel obligate intracellular gamma-proteobacterium associated with ixodid ticks, Diplorickettsia massiliensis, gen. nov., sp. nov. PLoS One 5:e11478. doi:10.1371/journal.pone.0011478
Kattar MM, Cookson BT, Carlson LDC, Stiglich SK, Schwartz MA, Nguyen TT, Daza R, Wallis CK, Yarfitz SL, Coyle MB (2001) Tsukamurella strandjordae sp. nov., a proposed new species causing sepsis. J Clin Microbiol 39:1467–1476. doi:10.1128/JCM.39.4.1467-1476.2001
Almehmi A, Pfister AK, McCowan R, Matulis S (2004) Implantable cardioverter-defibrillator infection caused by Tsukamurella. W V Med J 100:185–186
Kucerova Z, Stejskal V (2009) Morphological diagnosis of the eggs of stored-products mites. Exp Appl Acarol 49:173–183. doi:10.1007/s10493-009-9256-0
Ma W-J, Vavre F, Beukeboom LW (2014) Manipulation of arthropod sex determination by endosymbionts: diversity and molecular mechanisms. Sex Dev 8:59–73. doi:10.1159/000357024
Pietri JE, DeBruhl H, Sullivan W (2016) The rich somatic life of Wolbachia. MicrobiologyOpen 5:923–936. doi:10.1002/mbo3.390
Chirico J, Eriksson H, Fossum O, Jansson D (2003) The poultry red mite, Dermanyssus gallinae, a potential vector of Erysipelothrix rhusiopathiae causing erysipelas in hens. Med Vet Entomol 17:232–234. doi:10.1046/j.1365-2915.2003.00428.x
Eriksson H, Brannstrom S, Skarin H, Chirico J (2010) Characterization of Erysipelothrix rhusiopathiae isolates from laying hens and poultry red mites (Dermanyssus gallinae) from an outbreak of erysipelas. Avian Pathol 39:505–509. doi:10.1080/03079457.2010.518313
Acknowledgements
This study was supported by the project of the Ministry of Agriculture of the Czech Republic RO0417 and by COST Action FA1404 (COREMI) (http://www.coremi.eu/home.html). Computational resources were supplied by the Ministry of Education, Youth and Sports of the Czech Republic under the Projects CESNET (Project No. LM2015042) and CERIT-Scientific Cloud (Project No. LM2015085) provided within the program Projects of Large Research, Development and Innovations Infrastructures. We thank Vlastislav Machandr for his kind help with sample collection and Martin Markovic for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 2144 kb).
Rights and permissions
About this article
Cite this article
Hubert, J., Erban, T., Kopecky, J. et al. Comparison of Microbiomes between Red Poultry Mite Populations (Dermanyssus gallinae): Predominance of Bartonella-like Bacteria. Microb Ecol 74, 947–960 (2017). https://doi.org/10.1007/s00248-017-0993-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-0993-z