Abstract
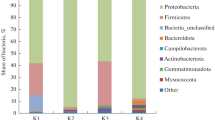
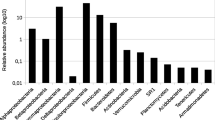
The long-term disposal of radioactive wastes in a deep geological repository is the accepted international solution for the treatment and management of these special residues. The microbial community of the selected host rocks and engineered barriers for the deep geological repository may affect the performance and the safety of the radioactive waste disposal. In this work, the bacterial population of bentonite formations of Almeria (Spain), selected as a reference material for bentonite-engineered barriers in the disposal of radioactive wastes, was studied. 16S ribosomal RNA (rRNA) gene-based approaches were used to study the bacterial community of the bentonite samples by traditional clone libraries and Illumina sequencing. Using both techniques, the bacterial diversity analysis revealed similar results, with phylotypes belonging to 14 different bacterial phyla: Acidobacteria, Actinobacteria, Armatimonadetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Deinococcus-Thermus, Firmicutes, Gemmatimonadetes, Planctomycetes, Proteobacteria, Nitrospirae, Verrucomicrobia and an unknown phylum. The dominant groups of the community were represented by Proteobacteria and Bacteroidetes. A high diversity was found in three of the studied samples. However, two samples were less diverse and dominated by Betaproteobacteria.






Similar content being viewed by others
References
Thury M, Bossart P (1999) The Mont Terri rock laboratory, a new international research project in a Mesozoic shale formation, in Switzerland. Eng Geol 52(3):347–359
Stroes-Gascoyne S, Hamon CJ, Dixon DA, Martino JB (2007) Microbial analysis of samples from the tunnel sealing experiment at AECL’s Underground Research Laboratory. Phys Chem Earth 32:219–231
Alonso EE, Springman SM, Ng CWW (2008) Monitoring large-scale tests for nuclear waste disposal. Geotech Geol Eng 26:817–826
Villar MV, Fernández-Soler JM, Delgado-Huertas A, Reyes E, Linares J, Jiménez de Cisneros C, Huertas FJ, Caballero E, Leguey S, Cuevas J, Garralón A, Fernández AM, Pelayo M, Martín PL, Pérez Del Villar L, Astudillo J (2006) The study of Spanish clays for their use as sealing materials in nuclear waste repositories: 20 years of progress. J Iber Geol 32:15–36
Stroes-Gascoyne S, Schippers A, Schwyn B, Poulain S, Sergeant C, Simonoff M, Le Marrec C, Altmann S, Nagaoka T, Mauclaire L, McKenzie J, Daumas S, Vinsot A, Beaucaire C, Matray JM (2007) Microbial community analysis of Opalinus Clay drill core samples from the Mont Terri Underground Research Laboratory, Switzerland. Geomicrobiol J 24:1–17
Poulain S, Sergeant C, Simonoff M, Le Marrec C, Altmann S (2008) Microbial investigations of Opalinus Clay, an argillaceous formation as a potential host rock under evaluation for a radioactive waste repository. Geomicrobiol J 25:240–249
Wouters K, Moors H, Boven P, Leys N (2013) Evidence and characteristics of a diverse and metabolically active microbial community in deep subsurface clay borehole water. FEMS Microbiol Ecol 86:458–473. doi:10.1111/1574-6941.12171
Cormenzana JL, García-Gutiérrez M, Missana T, Alonso U (2008) Modelling large-scale laboratory HTO and strontium diffusion experiments in Mont Terri and Bure clay rocks. Phys Chem Earth. doi:10.1016/j.pce.2008.05.006
Urios L, Marsal F, Pellegrini D, Magot M (2012) Microbial diversity of the 180 million-year-old Toarcian argillite from Tournemire, France. Appl Geochem 27:1442–1450. doi:10.1016/j.apgeochem.2011.09.022
Meleshyn A (2011) Microbial processes relevant for long-term performance of radioactive waste repositories in clays. GRS-291 Report. http://www.grs.de/publication/GRS-291
Macaskie LE, Bonthrone KM, Yong P, Goddard DT (2000) Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 146:1855–1867
Pedersen K (2002) Microbial processes in the disposal of high level radioactive waste 500 m underground in Fennoscandian Shield rocks. In: Keith-Roach MJ, Livens FR (ed) Interactions of microorganisms with radionuclides. Elsevier Science Ltd, pp 279–311
Merroun ML, Raff J, Rossberg A, Hennig C, Reich T, Selenska-Pobell S (2005) Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl Environ Microbiol 71:5532–5543
Lloyd JR, Renshaw JC (2005) Bioremediation of radioactive waste: radionuclide–microbe interactions in laboratory and field-scale studies. Curr Opin Biotechnol 16:254–260
Merroun ML, Nedelkova M, Ojeda JJ, Reitz T, López-Fernández M, Arias JM, Romero-González M, Selenska-Pobell S (2011) Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X-ray absorption spectroscopic analyses. J Hazard Mater 197:1–10
Lütke L, Moll H, Bachvarova V, Selenska-Pobell S, Bernhard G (2013) The U(VI) speciation influenced by a novel Paenibacillus isolate from Mont Terri Opalinus clay. Dalton Trans 42(19):6979–6988
Moll H, Lütke L, Bachvarova V, Cherkouk A, Selenska-Pobell S, Bernhard G (2014) Interactions of the Mont Terri Opalinus Clay isolate Sporomusa sp. MT-2.99 with curium(III) and europium(III). Geomicrobiol J. doi:10.1080/01490451.2014.889975
West JM, Mckinley IG, Chapman NA (1982) Microbes in deep geological systems and their possible influence on radioactive waste disposal. Radioact Waste Manage Nuclear Fuel Cycle 3:1–15
Pentrakova L, Su K, Pentrak M, Stucki JW (2013) A review of microbial redox interactions with structural Fe in clay minerals. Clay Miner 48:543–560
Glass JB, Orphan VJ (2012) Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front Microbiol 3:61
Stroes-Gascoyne S, Sergeant C, Schippers A, Hamon CJ, Nèble S, Vesvres MH, Barsotti V, Poulain S, Le Marrec C (2011) Biogeochemical processes in a clay formation in situ experiment: part D—microbial analyses—synthesis of results. Appl Geochem 26:980–989
Lopez-Fernandez M, Fernandez-Sanfrancisco O, Moreno-Garcia A, Martin-Sanchez I, Sanchez-Castro I, Merroun ML (2014) Microbial communities in bentonite formations and their interactions with uranium. Appl Geochem 49:77–86. doi:10.1016/j.apgeochem.2014.06.022
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740
Service RF (1997) Microbiologists explore life’s rich, hidden kingdoms. Science 275:1740–1742
Cardenas E, Tiedje JM (2008) New tools for discovering and characterizing microbial diversity. Curr Opin Biotechnol 19:544–549
Degnan PH, Ochman H (2012) Illumina-based analysis of microbial community diversity. ISMEJ 6:183–19427
Caballero E, Jimenez De Cisneros C, Huertas FJ, Huertas F, Pozzuoli A, Linares J (2005) Bentonites from Cabo de Gata, Almeria, Spain: a mineralogical and geochemical overview. Clay Miner 40:463–480
Martínez JA, Jiménez de Cisneros C, Caballero E (2007) Natural acid sulphate alteration in bentonites (Cabo de Gata, Almeria, SE Spain). Clay Miner 42(1):89–107
de Jongh WK (1973) X-ray fluorescence analysis applying theoretical matrix corrections. Stainless steel. X-Ray Spectrom 2:151–158. doi:10.1002/xrs.1300020404
Walkley A, Black IA (1934) An examination of Degtjareff method for determining soil organic matter and a proposed modification of the chromic acid titration method. Soil Sci 37:29–37
Mingorance MD, Barahona E, Fernandez-Galvez J (2007) Guidelines for improving organic carbon recovery by the wet oxidation method. Chemosphere 68:409–413
Selenska-Pobell S, Kampf G, Flemming K, Radeva G, Satchanska G (2001) Bacterial diversity in soil samples from two uranium waste piles as determined by rep-APD, RISA and 16S rDNA retrieval. Antonie Van Leeuwenhoek 79:149–161
Plassart P, Terrat S, Thomson B, Griffiths R, Dequiedt S, Lelievre M, Regnier T, Nowak V, Bailey M, Lemanceau P, Bispo A, Chabbi A, Maron PA, Mougel C, Ranjard L (2012) Evaluation of the ISO Standard 11063 DNA extraction procedure for assessing soil microbial abundance and community structure. PLoS One 7(9):e44279. doi:10.1371/journal.pone.0044279
Selenska-Pobell S (2002) Diversity and activity of bacteria in uranium waste piles. In: Keith-Roach MJ, Livens FR (ed) Interactions of microorganisms with radionuclides. Elsevier Science Ltd, pp 225–254
Camarinha-Silva A, Jauregui R, Chaves-Moreno D, Oxley APA, Schaumburg F, Becker K, Wos-Oxley ML, Pieper DH (2014) Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol. doi:10.1111/1462-2920.12362
Bohorquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, Kolter R, Junca H, Baena S, Zambrano MM (2012) In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol 63:103–115. doi:10.1007/s00248-011-9943-3
Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols, 2010(6), pdb-prot5448
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R (2008) Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Turner S, Pryer KM, Miao VPW, Palmer JD (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46:327–338
Geissler A, Selenska-Pobell S (2005) Addition of U(VI) to a uranium mining waste sample and resulting changes in the indigenous bacterial community. Geobiology 3:275–285
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
R Development Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Linares J, Huertas F, Lachica M, Reyes E (1973) Geochemistry of trace elements during the genesis of coloured bentonites. In: Serratosa JM (ed) Proceedings of the international clay conference 1972. CSIC, Madrid, pp 351–360
Delgado A (1993) Estudio isotopico de los procesos diageneticos e hidrotermales relacionados con la genesis de bentonitas (Cabo de Gata, Almeria). Doctoral Thesis, University of Granada, Spain
Caballero E (1985) Quimismo del proceso de bentonitización en la región volcánica de Cabo de Gata (Almería). Doctoral Thesis, University of Granada, Spain
Marcos N (2004) Results of the studies on bentonite samples from Serrata de Nijar, Almeria, Spain. Working Report 2004–24, Helsinki University of Technology
Garcia-Romero E (2012) Bentonitas del sureste de la Península Ibérica. Workshop Guia de Campo. Sociedad española de arcillas. ISBN: 978-84-695-3858-6
Reyes E, Caballero E, Huertas F, Linares J (1987) Bentonite deposits from Cabo de Gata region, Almería, Spain. Field Book Guide, Euroclay-87, Excursion A, 9–32
Nagra (2002) Project Opalinus Clay—safety report: demonstration of disposal feasibility for spent fuel, vitrified high-level waste and long-lived intermediate level waste (Entsorgungsnachweis). Nagra Technical Report NTB 02-05; Nagra: Switzerland
Van Geet M, Maes N, Dierckx A (2003) Characteristics of the Boom Clay organic matter, a review. Geological Survey of Belgium, Professional paper 2003/1, N. 298: 1–23
Yoon MH, Im WT (2007) Flavisolibacter ginsengiterrae gen. nov., sp. nov. and Flavisolibacter ginsengisoli sp. nov., isolated from ginseng cultivating soil. Int J Syst Evol Microbiol 57(8):1834–1839
Swanner ED, Nell RM, Templeton AS (2011) Ralstonia species mediate Fe-oxidation in circum neutral, metal-rich subsurface fluids of Henderson mine, CO. Chem Geol 284:339–350
Utturkar SM, Bollmann A, Brzoska RM, Klingeman DM, Epstein SE, Palumbo AV, Brown SD (2013) Draft genome sequence for Ralstonia sp. strain OR214, a bacterium with potential for bioremediation. Genome Announc 1(3):e00321-13. doi:10.1128/genomeA.00321-13
Mijnendonckx K, Provoost A, Ott CM, Venkateswaran K, Mahillon J, Leys N, Van Houdt R (2013) Characterization of the survival ability of Cupriavidus metallidurans and Ralstonia pickettii from space-related environments. Microb Ecol 65(2):347–360
Borremans B, Hobman JL, Provoost A, Brown NL, van Der Lelie D (2001) Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol 183:5651–5658
Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Médigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M (2010) The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5(5):e10433. doi:10.1371/journal.pone.0010433
Piotrowska-Seget Z, Cycon M, Kozdroj J (2005) Metal-tolerant bacteria occurring in heavily polluted soil and mine spoil. Appl Soil Ecol 28:237–246
Majumder A, Bhattacharyya K, Bhattacharyya S, Kole SC (2013) Arsenic-tolerant, arsenite-oxidising bacterial strains in the contaminated soils of West Bengal, India. Sci Total Environ 463:1006–1014
Urios L, Marsal F, Pellegrini D, Magot M (2013) Microbial diversity at iron-clay interfaces after 10 years of interaction inside a deep argillite geological formation (Tournemire, France). Geomicrobiol J 30(5):442–453
Onyenwoke RU, Brill JA, Farahi K, Wiegel J (2004) Sporulation genes in members of the low G + C Gram-type-positive phylogenetic branch (Firmicutes). Arch Microbiol 182:182–192
Stroes-Gascoyne S, West JM (1997) Microbial studies in the Canadian nuclear fuel waste management program. FEMS Microbiol Rev 20(3–4):573–590
Smart NR, Rance AP, Reddy B, Hallbeck L, Pedersen K, Johansson AJ (2014) In situ evaluation of model copper-cast iron canisters for spent nuclear fuel: a case of microbiologically influenced corrosion (MIC). Corros Eng Sci Technol 49(6):548–553
Hedrich S, Schlömann M, Johnson MD (2011) The iron-oxidizing Proteobacteria. Microbiology 157:1551–1564
Münzinger M, Taraz K, Budzikiewicz H, Drechsel H, Heymann P, Winkelmann G, Meyer JM (1999) S, S-rhizoferrin (enantio-rhizoferrin)—a siderophore of Ralstonia (Pseudomonas) pickettii DSM 6297—the optical antipode of R, R-rhizoferrin isolated from fungi. Biometals 12:189–193
Shelobolina E, Konishi H, Xu H, Benzine J, Xiong MY, Wu T, Blöthe M, Roden E (2012) Isolation of phyllosilicate–iron redox cycling microorganisms from an illite-smectite rich hydromorphic soil. Front Microbiol 3:134–143
Sun H, Shi B, Bai Y, Wang D (2014) Bacterial community of biofilms developed under different water supply conditions in a distribution system. Sci Total Environ 472:99–107
Li D, Li Z, Yu J, Cao N, Liu R, Yang M (2010) Characterization of bacterial community structure in a drinking water distribution system during an occurrence of red water. Appl Environ Microbiol 76:7171–7180
Acknowledgments
This work was funded by the ERDF-cofinanced Grants CGL2012-36505 (80 % finding by FEDER) and CGL2009-09760 (70 % finding by FEDER) as well as BES-2010-032098 and EEBB-2011-43985 (Ministerio de Ciencia e Innovación, Spain). We acknowledge the assistance of F. Javier Huertas-Puerta (Instituto Andaluz de Ciencias de la Tierra, CSIC–University of Granada, Spain) with the XRD analyses; of Jose Antonio Rodriguez-Liebana (Instituto Andaluz de Ciencias de la Tierra, CSIC, Granada, Spain) with the TOC content determination; of Monika Dudek and Katrin Flemming (HZDR, Germany) with the clone libraries sequencing; of Gert Bernhard (HZDR, Germany) for his support; and of Iris Plumeier (HZI, Germany) with the Illumina sequencing.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Online Resource 1
(DOCX 196 kb)
Online Resource 2
(DOCX 22 kb)
Online Resource 3
(DOCX 293 kb)
Online Resource 4
(DOCX 45 kb)
Online Resource 5
(DOCX 26 kb)
Online Resource 6
(DOCX 30 kb)
Online Resource 7
(DOCX 28 kb)
Online Resource 8
(DOCX 124 kb)
Online Resource 9
(DOCX 18 kb)
Online Resource 10
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Lopez-Fernandez, M., Cherkouk, A., Vilchez-Vargas, R. et al. Bacterial Diversity in Bentonites, Engineered Barrier for Deep Geological Disposal of Radioactive Wastes. Microb Ecol 70, 922–935 (2015). https://doi.org/10.1007/s00248-015-0630-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0630-7