Abstract
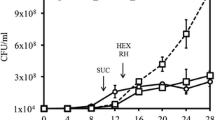
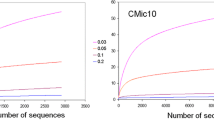
Red clay is a type of soil, the red color of which results from the presence of iron oxide. It is considered an eco-friendly material, with many industrial, cosmetic, and architectural uses. A patented method was applied to red clay in order to change its chemical composition and mineral bioavailability. The resulting product was designated processed red clay. This study evaluates the novel use of red clay and processed red clay as biostimulation agents in diesel-contaminated soils. Diesel biodegradation was enhanced in the presence of red clay and processed red clay by 4.9- and 6.7-fold, respectively, and the number of culturable bacterial cells was correlated with the amount of diesel biodegradation. The growth of Acinetobacter oleivorans DR1, Pseudomonas putida KT2440, and Cupriavidus necator was promoted by both types of red clays. Culture-independent community analysis determined via barcoded pyrosequencing indicated that Nocardioidaceae, Xanthomonadaceae, Pseudomonadaceae, and Caulobacteraceae were enriched by diesel contamination. Bacterial strain isolation from naphthalene- and liquid paraffin-amended media was affiliated with enriched taxa based on 16S rRNA gene sequence identity. We suggest that the biostimulating mechanism of red clay and processed red clay is able to support bacterial growth without apparent selection for specific bacterial species.





Similar content being viewed by others
References
Tyagi M, da Fonseca MM, de Carvalho CC (2011) Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 22:234–241
Bartha R (1986) Biotechnology of petroleum pollutant biodegradation. Microb Ecol 12:155–172
Swannell RP, Lee K, McDonagh M (1996) Field evaluations of marine oil spill bioremediation. Microbiol Rev 60:342–365
Balba MT, Al-Awadhi N, Al-Daher R (1998) Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J Microbiol Methods 32:155–164
Jimenez N, Vinas M, Sabate J, Diez S, Bayona JM, Solanas AM, Albaiges J (2006) The Prestige oil spill. 2. Enhanced biodegradation of a heavy fuel oil under field conditions by the use of an oleophilic fertilizer. Environ Sci Technol 40:2578–2585
Delille D, Pelletier E, Rodriguez-Blanco A, Ghiglione JF (2009) Effects of nutrient and temperature on degradation of petroleum hydrocarbons in sub-Antarctic coastal seawater. Polar Biol 32:1521–1528
Atlas RM (1995) Bioremediation of petroleum pollutants. Int Biodeterior Biodegrad 35:317–327
Bordoloi NK, Konwar BK (2009) Bacterial biosurfactant in enhancing solubility and metabolism of petroleum hydrocarbons. J Hazard Mater 170:495–505
Nikolopoulou M, Pasadakis N, Norf H, Kalogerakis N (2013) Enhanced ex situ bioremediation of crude oil contaminated beach sand by supplementation with nutrients and rhamnolipids. Mar Pollut Bull. doi:10.1016/j.marpolbul.2013.10.038
Nikolopoulou M, Kalogerakis N (2008) Enhanced bioremediation of crude oil utilizing lipophilic fertilizers combined with biosurfactants and molasses. Mar Pollut Bull 56:1855–1861
Silva-Castro GA, Rodelas B, Perucha C, Laguna J, González-López J, Calvo C (2013) Bioremediation of diesel-polluted soil using biostimulation as post-treatment after oxidation with Fenton-like reagents: assays in a pilot plant. Sci Total Environ 445–446:347–355
Shukor MY, Dahalan FA, Jusoh AZ, Muse R, Shamaan NA, Syed MA (2009) Characterization of a diesel-degrading strain isolated from a hydrocarbon-contaminated site. J Environ Biol 30:145–150
Jung J, Baek JH, Park W (2010) Complete genome sequence of the diesel-degrading Acinetobacter sp. strain DR1. J Bacteriol 192:4794–4795
Powell SM, Ferguson SH, Bowman JP, Snape I (2006) Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb Ecol 52:523–532
Salminen JM, Tuomi PM, Jorgensen KS (2008) Functional gene abundances (nahAc, alkB, xylE) in the assessment of the efficacy of bioremediation. Appl Biochem Biotechnol 151:638–652
Yergeau E, Arbour M, Brousseau R, Juck D, Lawrence JR, Masson L, Whyte LG, Greer CW (2009) Microarray and real-time PCR analyses of the responses of high Arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl Environ Microbiol 75:6258–6267
Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol 69:1800–1809
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000
Jurelevicius D, Alvarez VM, Marques JM, de Sousa Lima LR, Dias Fde A, Seldin L (2013) Bacterial community response to petroleum hydrocarbon amendments in freshwater, marine, and hypersaline water-containing microcosm. Appl Environ Microbiol 79:5927–5935
Jung J, Madsen EL, Jeon CO, Park W (2011) Comparative genomic analysis of Acinetobacter oleivorans DR1 to determine strain-specific genomic regions and gentisate biodegradation. Appl Environ Microbiol 77:7418–7424
Hwang JY, Jang MI, Kim JS, Cho WM, Ahn BS, Kang SW (2000) Mineralogy and chemical composition of the residual soils (Hwangto) from south Korea. J Miner Soc Korea 13:147–163
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Jeraldo P, Chia N, Goldenfeld N (2011) On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol 13:3000–3009
Warr LN, Friese A, Schwarz F, Schauer F, Portier RJ, Basirico LM, Olson GM (2013) Bioremediating oil spills in nutrient poor ocean waters using fertilized clay mineral flakes: some experimental constraints. Biotechnol Res Int 2013:704806
Sarkar D, Ferguson M, Datta R, Birnbaum S (2005) Bioremediation of petroleum hydrocarbons in contaminated soils: comparison of biosolids addition, carbon supplementation, and monitored natural attenuation. Environ Pollut 136:187–195
da Silva AC, de Oliveira FJ, Bernardes DS, de França FP (2009) Bioremediation of marine sediments impacted by petroleum. Appl Biochem Biotechnol 153:58–66
Coulon F, Brassington KJ, Bazin R, Linnet PE, Thomas KA, Mitchell TR, Lethbridge G, Smith JW, Pollarda SJ (2012) Effect of fertilizer formulation and bioaugmentation on biodegradation and leaching of crude oils and refined products in soils. Environ Technol 33:1879–1893
Ding GC, Heuer H, Smalla K (2012) Dynamics of bacterial communities in two unpolluted soils after spiking with phenanthrene: soil type specific and common responders. Front Microbiol 3:290
Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW (2013) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. IJME J 7:1200–1210
Sutton NB, Maphosa F, Morillo JA, Abu Al-Soud W, Langenhoff AA, Grotenhuis T, Rijnaarts HH, Smidt H (2013) Impact of long-term diesel contamination on soil microbial community structure. Appl Environ Microbiol 79:619–630
Acknowledgement
This work was supported by a grant (Grant# 812001-3) from the Institute of Planning and Evaluation for Technology of Agriculture, Forestry, Fisheries and Food (IPET, Republic of Korea).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jung, J., Choi, S., Hong, H. et al. Effect of Red Clay on Diesel Bioremediation and Soil Bacterial Community. Microb Ecol 68, 314–323 (2014). https://doi.org/10.1007/s00248-014-0420-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0420-7