Abstract
Marine bacteria from aquaculture areas with industrial use of quinolones have the potential to pass quinolone resistance genes to animal and human pathogens. The VPA0095 gene, related to the quinolone resistance determinant qnrA, from clinical isolates of epidemic Vibrio parahaemolyticus conferred reduced susceptibility to quinolone after cloning into Escherichia coli K-12 either when acting alone or synergistically with DNA gyrase mutations. In addition, a plasmid-mediated quinolone resistance gene from marine bacteria, aac(6′)-Ib-cr, was identical to aac(6′)-Ib-cr from urinary tract isolates of E. coli, suggesting a recent flow of this gene between these bacteria isolated from different environments. aac(6′)-Ib-cr from E. coli also conferred reduced susceptibility to quinolone and kanamycin when cloned into E. coli K-12.

Similar content being viewed by others
References
Berge AC, Atwill ER, Sischo WM (2005) Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev Vet Med 69:25–38
Prescott JF (2006) History of antimicrobial usage in agriculture. In: Aarestrup FM (ed) Antimicrobial resistance in bacteria of animal origin, 1st edn. ASM Press, Washington, pp 19–27
Cox LA Jr, Ricci PF (2008) Causal regulations vs. political will: why human zoonotic infections increase despite precautionary bans on animal antibiotics. Environ Int 34:459–475
Marshall BM, Levy SB (2011) Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 24:718–733
Phillips I, Casewell M, Cox T, De Groot B, Friis C, Jones R, Nightingale C, Preston R, Waddell J (2004) Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother 53:28–52
Kloos WE, Ballard DN, Webster JA, Hubner RJ, Tomasz A, Couto I, Sloan GL, Dehart HP, Fiedler F, Schubert K, de Lencastre H, Sanches IS, Heath HE, Leblanc PA, Ljungh A (1997) Ribotype delineation and description of Staphylococcus sciuri subspecies and their potential as reservoirs of methicillin resistance and staphylolytic enzyme genes. Int J Syst Bacteriol 47:313–323
Wu S, Piscitelli C, de Lencastre H, Tomasz A (1996) Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist 2:435–441
Klare I, Heier H, Claus H, Bohme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W (1995) Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist 1:265–272
Bates J, Jordens Z, Selkon JB (1993) Evidence for an animal origin of vancomycin-resistant enterococci. Lancet 342:490–491
Molbak K (2005) Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infect Dis 41:1613–1620
Heuer OE, Kruse H, Grave K, Collignon P, Karunasagar I, Angulo FJ (2009) Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis 49:1248–1253
Buschmann AH, Tomova A, Lopez A, Maldonado MA, Henriquez LA, Ivanova L, Moy F, Godfrey HP, Cabello FC (2012) Salmon aquaculture and antimicrobial resistance in the marine environment. PLoS One 7:e42724
Sapkota A, Sapkota AR, Kucharski M, Burke J, McKenzie S, Walker P, Lawrence R (2008) Aquaculture practices and potential human health risks: current knowledge and future priorities. Environ Int 34:1215–1226
Cabello FC, Godfrey HP, Tomova A, Ivanova L, Dolz H, Millanao A, Buschmann AH (2013) Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol 15:1917–1942
Saga T, Kaku M, Onodera Y, Yamachika S, Sato K, Takase H (2005) Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob Agents Chemother 49:2144–2145
Poirel L, Liard A, Rodriguez-Martinez JM, Nordmann P (2005) Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother 56:1118–1121
Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P (2005) Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother 49:3523–3525
Martinez-Martinez L, Pascual A, Jacoby GA (1998) Quinolone resistance from a transferable plasmid. Lancet 351:797–799
Nordmann P, Poirel L (2005) Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother 56:463–469
Cambau E, Lascols C, Sougakoff W, Bebear C, Bonnet R, Cavallo JD, Gutmann L, Ploy MC, Jarlier V, Soussy CJ, Robert J (2006) Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002–2005. Clin Microbiol Infect 12:1013–1020
Gonzalez-Escalona N, Cachicas V, Acevedo C, Rioseco ML, Vergara JA, Cabello F, Romero J, Espejo RT (2005) Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg Infect Dis 11:129–131
Wang H, Dzink-Fox JL, Chen M, Levy SB (2001) Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45:1515–1521
Bouanchaud DH, Scavizzi MR, Chabbert YA (1968) Elimination by ethidium bromide of antibiotic resistance in enterobacteria and staphylococci. J Gen Microbiol 54:417–425
Yoshida H, Bogaki M, Nakamura M, Nakamura S (1990) Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother 34:1271–1272
Dridi L, Tankovic J, Burghoffer B, Barbut F, Petit JC (2002) gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob Agents Chemother 46:3418–3421
Zaiss NH, Witte W, Nubel U (2010) Fluoroquinolone resistance and Clostridium difficile, Germany. Emerg Infect Dis 16:675–677
Chen DQ, Yang L, Luo YT, Mao MJ, Lin YP, Wu AW (2013) Prevalence and characterization of quinolone resistance in Laribacter hongkongensis from grass carp and Chinese tiger frog. J Med Microbiol 62:1559–1564
Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC (2006) Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88
Martinez JL, Baquero F (2000) Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44:1771–1777
Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A (2009) Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 22:664–689
Acknowledgments
This work was supported by a grant from the Lenfest Ocean Program/Pew Charitable Trusts to F.C.C. and by a fellowship from the John Simon Guggenheim Foundation to F.C.C. that allowed him to begin to study antimicrobial use in aquaculture. We thank Dr. Romilio Espejo, Universidad de Chile, Santiago, Chile for V. parahaemolyticus strains. We thank Dr. Maria L. Rioseco, Hospital Regional de Puerto Montt, Chile, for the E. coli clinical isolates used in this study. Dr. D.C. Hooper, Massachusetts General Hospital, Boston, MA, USA, for J53AzR. We also thank Dr. Henry P. Godfrey for his important help for improving the text and Mrs. Harriett Harrison for preparation of the manuscript. We thank Mariya Sambir and Rene Devis for assistance with some experiments.
Authors’ contributions
SA identified the genes VPA0095 and aac(6′)-Ib-cr in V. parahaemolyticus and E. coli clinical isolates, respectively, cloned the genes, performed susceptibility tests, isolated the chromosomal mutant resistant to quinolones, cured plasmid, carried out DNA sequence analysis, and participated in manuscript drafting. LI and AT kept and studied antimicrobial susceptibility of marine bacteria, sequenced their aac(6′)-Ib-cr, and performed their DNA sequence analysis. FC obtained the funds for this work, planned the experiments, analyzed the data, and participated in manuscript drafting.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Fig. S1
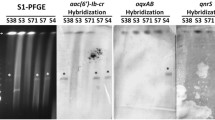
Detection by DNA hybridization of aac(6′)-Ib-cr gene in plasmid DNA from urinary tract isolates of E. coli resistant to quinolones. A. Plasmid DNA from E. coli clinical isolates. B. Plasmid DNA hybridization with a aac(6′)-Ib-cr probe. Lanes: 1. isolate 165; 2. isolate 110; 3. isolate 248; 4. isolate 435; 5. isolate 562; 6. isolate 146; 7. isolate 189; 8. isolate 580; 9. isolate 207; 10. isolate 204; 11. E. coli DH5α pUC19-aac(6′)-Ib-cr (positive control); 12. pBR328 (negative control) (PPTX 74 kb)
Table S1
(DOCX 11 kb)
Rights and permissions
About this article
Cite this article
Aedo, S., Ivanova, L., Tomova, A. et al. Plasmid-Related Quinolone Resistance Determinants in Epidemic Vibrio parahaemolyticus, Uropathogenic Escherichia coli, and Marine Bacteria from an Aquaculture Area in Chile. Microb Ecol 68, 324–328 (2014). https://doi.org/10.1007/s00248-014-0409-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0409-2