Abstract
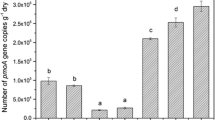
Little is understood about the relationship between microbial assemblage history, the composition and function of specific functional guilds and the ecosystem functions they provide. To learn more about this relationship we used methane oxidizing bacteria (MOB) as model organisms and performed soil microcosm experiments comprised of identical soil substrates, hosting distinct overall microbial diversities (i.e., full, reduced and zero total microbial and MOB diversities). After inoculation with undisturbed soil, the recovery of MOB activity, MOB diversity and total bacterial diversity were followed over 3 months by methane oxidation potential measurements and analyses targeting pmoA and 16S rRNA genes. Measurement of methane oxidation potential demonstrated different recovery rates across the different treatments. Despite different starting microbial diversities, the recovery and succession of the MOB communities followed a similar pattern across the different treatment microcosms. In this study we found that edaphic parameters were the dominant factor shaping microbial communities over time and that the starting microbial community played only a minor role in shaping MOB microbial community




Similar content being viewed by others
References
Torsvik V, Øvreås L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245
Bodelier PL (2011) Toward understanding, managing, and protecting microbial ecosystems. Front Microbiol 2:80. doi:10.3389/fmicb.2011.00080
Conrad R, Klose M, Noll M (2009) Functional and structural response of the methanogenic microbial community in rice field soil to temperature change. Environ Microbiol 11:1844–1853
Conrad R, Donald LS (2007) Microbial ecology of methanogens and methanotrophs advances in agronomy. Academic Press, pp. 1–63
Semrau JD, DiSpirito AA, Yoon S (2010) Methanotrophs and copper. FEMS Microbiol Rev 34:496–531
Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M (2007) Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882
Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K (2008) Methane oxidation at 55 °C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci U S A 105:300–304. doi:10.1073/pnas.0704162105
McDonald IR, Bodrossy L, Chen Y, Murrell JC (2008) Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol 74:1305–1315
Ho A, Kerckhof FM, Luke C, Reim A, Krause S, Boon N, Bodelier PLE (2013) Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ Microbiol Rep 5:335–345
Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J (2008) Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62:375–401
Chase JM (2003) Community assembly: when should history matter? Oecologia 136:489–498
Chase JM (2010) Stochastic community assembly causes higher biodiversity in more Productive Environments. Science 328:1388–1391. doi:10.1126/science.1187820
Caruso T, Chan Y, Lacap DC, Lau MCY, McKay CP, Pointing SB (2011) Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J 5:1406–1413
Langenheder S, Szekely AJ (2011) Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5:1086–1094
Fukami T, Morin PJ (2003) Productivity–biodiversity relationships depend on the history of community assembly. Nature 424:423–426
Cardinale BJ (2011) Biodiversity improves water quality through niche partitioning. Nature 472:86–89
Langenheder S, Lindsrt ES, Tranvik LJ (2005) Weak coupling between community composition and functioning of aquatic bacteria. American Society of Limnology and Oceanography. Waco, TX, ETATS-UNIS
Degens BP, Schipper LA, Sparling GP, Duncan LC (2001) Is the microbial community in a soil with reduced catabolic diversity less resistant to stress or disturbance? Soil Biol Biochem 33:1143–1153
Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, De Vos P, Verstraete W, Boon N (2009) Initial community evenness favours functionality under selective stress. Nature 458:623–626
Bressan M, Mougel C, Dequiedt S, Maron P-A, Lemanceau P, Ranjard L (2008) Response of soil bacterial community structure to successive perturbations of different types and intensities. Environ Microbiol 10:2184–2187
Mohanty SR, Bodelier PLE, Floris V, Conrad R (2006) Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl Environ Microbiol 72:1346–1354
Shade A, Peter H, Allison SD, Baho DL, Berga M, Burgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JB, Matulich KL, Schmidt TM, Handelsman J (2012) Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417
Kemnitz D, Chin K-J, Bodelier P, Conrad R (2004) Community analysis of methanogenic archaea within a riparian flooding gradient. Environ Microbiol 6:449–461
Bodelier PL, Bar-Gilissen MJ, Meima-Franke M, Hordijk K (2012) Structural and functional response of methane-consuming microbial communities to different flooding regimes in riparian soils. Ecol Evol 2:106–127
Steenbergh AK, Meima MM, Kamst M, Bodelier PLE (2010) Biphasic kinetics of a methanotrophic community is a combination of growth and increased activity per cell. FEMS Microbiol Ecol 71:12–22
Bodrossy L, Stralis-Pavese N, Murrell JC, Radajewski S, Weilharter A, Sessitsch A (2003) Development and validation of a diagnostic microbial microarray for methanotrophs. Environ Microbiol 5:566–582
Kolb S, Knief C, Stubner S, Conrad R (2003) Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl Environ Microbiol 69:2423–2429
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
Dunbar J, Ticknor LO, Kuske CR (2001) Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol 67:190–197
Abell GCJ, Stralis-Pavese N, Sessitsch A, Bodrossy L (2009) Grazing affects methanotroph activity and diversity in an alpine meadow soil. Environ Microbiol Rep 1:457–465
Anderson MJ, Gorley RN, Clarke KR (2008) PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. PRIMER-E, Plymouth, UK
Wang J, Krause S, Muyzer G, Meima-Franke M, Laanbroek HJ, Bodelier PL (2012) Spatial patterns of iron- and methane-oxidizing bacterial communities in an irregularly flooded, riparian wetland. Front Microbiol 3:64
Bodelier PLE, Frenzel P (1999) Contribution of methanotrophic and nitrifying bacteria to CH4 and NH4 + oxidation in the rhizosphere of rice plants as determined by new methods of discrimination. Appl Environ Microbiol 65:1826–1833
Heifets L, Lindholm-Levy P (1989) Comparison of bactericidal activities of streptomycin, amikacin, kanamycin, and capreomycin against Mycobacterium avium and M. tuberculosis. Antimicrob Agents Chemother 33:1298–1301
Hidding B, Nolet BA, De Boer T, De Vries PP, Klaassen M (2009) Compensatory growth in an aquatic plant mediates exploitative competition between seasonally tied herbivores. Ecology 90:1891–1899
Ho A, Luke C, Frenzel P (2011) Recovery of methanotrophs from disturbance: population dynamics, evenness and functioning. ISME J 5:750–758
Eller G, Frenzel P (2001) Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl Environ Microbiol 67:2395–2403
Gebert J, Singh BK, Pan Y, Bodrossy L (2009) Activity and structure of methanotrophic communities in landfill cover soils. Environ Microbiol Rep 1:414–423
Siljanen HMP, Saari A, Krause S, Lensu A, Abell GCJ, Bodrossy L, Bodelier PLE, Martikainen PJ (2011) Hydrology is reflected in the functioning and community composition of methanotrophs in the littoral wetland of a boreal lake. FEMS Microbiol Ecol 75:430–445
Anderson MJ, Ellingsen KE, McArdle BH (2006) Multivariate dispersion as a measure of beta diversity. Ecol Lett 9:683–693
Chase JM, Leibold MA (2002) Spatial scale dictates the productivity–biodiversity relationship. Nature 416:427–430
Langenheder S, Berga M, Ostman O, Szekely AJ (2011) Temporal variation of beta-diversity and assembly mechanisms in a bacterial metacommunity. ISME J 6:1107–1114
Belova SE, Kulichevskaya IS, Bodelier PLE, Dedysh SN (2013) Methylocystis bryophila sp nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int J Syst Evol Microbiol 63:1096–1104
Semrau JD, DiSpirito AA, Vuilleumier S (2011) Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett 323:1–12
Acknowledgements
We thank Zoltán Kerényi for assistance with microcosms. Research at AIT was supported by the ESF EuroDiversity programme METHECO (No. FP018, local funding agency: FWF, Austria, project number I40-B06). YP received a travel grant from the Marine Biogeochemistry Program of CSIRO Marine and Atmospheric Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pan, Y., Abell, G.C.J., Bodelier, P.L.E. et al. Remarkable Recovery and Colonization Behaviour of Methane Oxidizing Bacteria in Soil After Disturbance Is Controlled by Methane Source Only. Microb Ecol 68, 259–270 (2014). https://doi.org/10.1007/s00248-014-0402-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0402-9