Abstract
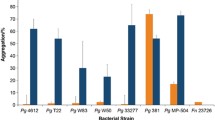
The oral opportunistic pathogen Fusobacterium nucleatum is known to interact with a large number of different bacterial species residing in the oral cavity. It adheres to a variety of Gram-positive bacteria, including oral streptococci via the arginine-inhibitable adhesin RadD. In this study, we describe a novel protein encoded by the predicted open reading frame FN1253 that appears to play a role in interspecies interactions of F. nucleatum, particularly with oral streptococci and related Gram-positive species. We designated FN1253 as aid1 (Adherence Inducing Determinant 1). Expression analyses demonstrated that this gene was induced in F. nucleatum single species biofilms, while the presence of representative members of the oral microbiota known to adhere to F. nucleatum triggered its suppression. Inactivation as well as overexpression of aid1 affected the ability of F. nucleatum to coaggregate with oral streptococci and the closely related Enterococcus faecalis, but not other Gram-positive oral species tested. Furthermore, overexpression of aid1 led to a drastic change in the structure of dual species biofilms of F. nucleatum with oral streptococci. Aid1 function was abolished in the presence of arginine and found to be dependent on RadD. Interestingly, differential expression of aid1 did not affect messenger RNA and protein levels of RadD. These findings indicate that RadD-mediated adhesion to oral streptococci involves more complex cellular processes than the simple interaction of adhesins on the surface of partner strains. Aid1 could potentially play an important role in facilitating RadD-mediated interaction with oral streptococci by increasing binding specificity of F. nucleatum to other microbial species.





Similar content being viewed by others
References
Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG (2010) The human oral microbiome. J Bacteriol 192(19):5002–5017. doi:10.1128/JB.00542-10
Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE (2005) Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43(11):5721–5732. doi:10.1128/JCM.43.11.5721-5732.2005
Kolenbrander PE, London J (1993) Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol 175(11):3247–3252
Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ (2002) Communication among oral bacteria. Microbiol Mol Biol Rev 66(3):486–505. doi:10.1128/mmbr.66.3.486-505.2002
Bastos JA, Diniz CG, Bastos MG, Vilela EM, Silva VL, Chaoubah A, Souza-Costa DC, Andrade LC (2011) Identification of periodontal pathogens and severity of periodontitis in patients with and without chronic kidney disease. Arch Oral Biol 56(8):804–811. doi:10.1016/j.archoralbio.2010.12.006
Gonzalez OA, Li M, Ebersole JL, Huang CB (2010) HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis involves toll-like receptor 2 [corrected] and 9 activation in monocytes/macrophages. CVI 17(9):1417–1427. doi:10.1128/CVI.00009-10
Hsiao WW, Li KL, Liu Z, Jones C, Fraser-Liggett CM, Fouad AF (2012) Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genomics 13:345. doi:10.1186/1471-2164-13-345
Lee HR, Jun HK, Kim HD, Lee SH, Choi BK (2012) Fusobacterium nucleatum GroEL induces risk factors of atherosclerosis in human microvascular endothelial cells and ApoE(−/−) mice. Mol Oral Microbiol 27(2):109–123. doi:10.1111/j.2041-1014.2011.00636.x
Okada M, Awane S, Suzuki J, Hino T, Takemoto T, Kurihara H, Miura K (2002) Microbiological, immunological and genetic factors in family members with periodontitis as a manifestation of systemic disease, associated with hematological disorders. J Periodontal Res 37(4):307–315
Sparks Stein P, Steffen MJ, Smith C, Jicha G, Ebersole JL, Abner E, Dawson D 3rd (2012) Serum antibodies to periodontal pathogens are a risk factor for Alzheimer’s disease. Alzheimers Dement JAlzheimer Assoc 8(3):196–203. doi:10.1016/j.jalz.2011.04.006
Han YW, Wang X (2013) Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res 92(6):485–491
Kaplan CW, Lux R, Haake SK, Shi W (2009) The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71(1):35–47. doi:10.1111/j.1365-2958.2008.06503.x
McHardy IH (2011) Analysis of interspecies interactions of periodontal bacteria using microarrays. Ph D, UCLA
Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D'Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R (2002) Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184(7):2005–2018
Ohta K, Makinen KK, Loesche WJ (1986) Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun 53(1):213–220
Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, Haake SK (2005) Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J Dent Res 84(8):700–704
Haake SK, Yoder SC, Attarian G, Podkaminer K (2000) Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J Bacteriol 182(4):1176–1180
Cisar JO, Kolenbrander PE, McIntire FC (1979) Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun 24(3):742–752
Kolenbrander PE, Andersen RN, Moore LV (1990) Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl Environ Microbiol 56(12):3890–3894
Kolenbrander PE, Andersen RN (1989) Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect Immun 57(10):3204–3209
Whittaker CJ, Klier CM, Kolenbrander PE (1996) Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol 50:513–552. doi:10.1146/annurev.micro.50.1.513
Takemoto T, Hino T, Yoshida M, Nakanishi K, Shirakawa M, Okamoto H (1995) Characteristics of multimodal co-aggregation between Fusobacterium nucleatum and streptococci. J Periodontal Res 30(4):252–257
ten Cate JM (2006) Biofilms, a new approach to the microbiology of dental plaque. Odontology 94(1):1–9. doi:10.1007/s10266-006-0063-3
Kolenbrander PE (1993) Coaggregation of human oral bacteria: potential role in the accretion of dental plaque. J Appl Bacteriol 74(Suppl):79S–86S
Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, Harmsen HJ (2010) Oral biofilm architecture on natural teeth. PLoS ONE 5(2):e9321. doi:10.1371/journal.pone.0009321
Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU (2005) Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci U S A 102(40):14278–14283. doi:10.1073/pnas.0501234102
Barwe SP, Kim S, Rajasekaran SA, Bowie JU, Rajasekaran AK (2007) Janus model of the Na, K-ATPase beta-subunit transmembrane domain: distinct faces mediate alpha/beta assembly and beta-beta homo-oligomerization. J Mol Biol 365(3):706–714. doi:10.1016/j.jmb.2006.10.029
Plotkowski ML, Kim S, Phillips ML, Partridge AW, Deber CM, Bowie JU (2007) Transmembrane domain of myelin protein zero can form dimers: possible implications for myelin construction. Biochemistry 46(43):12164–12173. doi:10.1021/bi701066h
Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX (2005) Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol 187(15):5330–5340
Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW (2007) FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem 282(34):25000–25009
Acknowledgments
The pHS58 shuttle plasmid was kindly provided by Dr. Susan Kinder Haake. This work is supported by the Whitcome Pre-doctoral Training Grant to AK and NIH/NIDCR grant DE021108 to RL.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Construction of aid1 mutant strains. A. Construction of Δaid1 inactivation strain. aid1 was inactivated via double homologous recombination by inserting a catP thiamphenicol/chloramphenicol resistance cassette into the aid1 gene. B. Construction of Fn/pEP-aid1 and radD/pEP-aid1 overexpression strains. aid1 was overexpressed in wild type F. nucleatum ATCC 23726 and in radD strains by introducing a shuttle plasmid carrying aid1 gene with its own endogenous promoter into wild type F. nucleatum ATCC 23726 and ΔradD strains respectively. (PPTX 52 kb)
Rights and permissions
About this article
Cite this article
Kaplan, A., Kaplan, C.W., He, X. et al. Characterization of aid1, a Novel Gene Involved in Fusobacterium nucleatum Interspecies Interactions. Microb Ecol 68, 379–387 (2014). https://doi.org/10.1007/s00248-014-0400-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0400-y