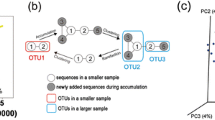
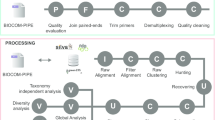
Abstract
Next-generation sequencing has increased the coverage of microbial diversity surveys by orders of magnitude, but differentiating artifacts from rare environmental sequences remains a challenge. Clustering 16S rRNA sequences into operational taxonomic units (OTUs) organizes sequence data into groups of 97 % identity, helping to reduce data volumes and avoid analyzing sequencing artifacts by grouping them with real sequences. Here, we analyze sequence abundance distributions across environmental samples and show that 16S rRNA sequences of >99 % identity can represent functionally distinct microorganisms, rendering OTU clustering problematic when the goal is an accurate analysis of organism distribution. Strict postsequencing quality control (QC) filters eliminated the most prevalent artifacts without clustering. Further experiments proved that DNA polymerase errors in polymerase chain reaction (PCR) generate a significant number of substitution errors, most of which pass QC filters. Based on our findings, we recommend minimizing the number of PCR cycles in DNA library preparation and applying strict postsequencing QC filters to reduce the most prevalent artifacts while maintaining a high level of accuracy in diversity estimates. We further recommend correlating rare and abundant sequences across environmental samples, rather than clustering into OTUs, to identify remaining sequence artifacts without losing the resolution afforded by high-throughput sequencing.





Similar content being viewed by others
References
Pace N (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740
Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA, Progra NCS (2009) Topographical and temporal diversity of the human skin microbiome. Science 324(5931):1190–1192
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103(32):12115–12120
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, Dewell SB, de Winter A, Drake J, Du L, Fierro JM, Forte R, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Hutchison SK, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lee WL, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Reifler M, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Willoughby DA, Yu PG, Begley RF, Rothberg JM (2006) Genome sequencing in microfabricated high-density picolitre reactors (vol 437, pg 376, 2005). Nature 441(7089):120–120
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1(4):283–290
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C (2009) Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3(7):860–869
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology 8(7):R143. doi:10.1186/gb-2007-8-7-r143
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12(1):118–123
Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6(9):639–U627
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12(38). doi:10.1186/1471-2105-12-38
von Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21(3):213–229
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW, Consortium HM (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21(3):494–504
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Engelbrektson A, Kunin V, Wrighton K, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P (2010) Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4:642–647
Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, Li HZ, Bushman FD (2010) Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. Bmc Microbiology 10(206). doi:10.1186/1471-2180-10-206
Cline J, Braman J, Hogrefe H (1996) PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res 24(18):3546–3551
Eckert KA, Kunkel TA (1991) DNA polymerase fidelity and the polymerase chain reaction. Genome Res 1(1):17–24
Huse SH, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12(7):1889–1898
Lundin D, Severin I, Logue JB, Ostman O, Andersson AF, Lindstrom ES (2012) Which sequencing depth is sufficient to describe patterns in bacterial alpha- and beta-diversity? Environ Microbiol Rep 4(3):367–372
Schloss PD (2010) The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. Plos Comput Biol 6(7):e1000844. doi:10.1371/journal.pcbi.1000844
Sipos M, Jeraldo P, Chia N, Qu AI, Dhillon AS, Konkel ME, Nelson KE, White BA, Goldenfeld N (2010) Robust computational analysis of rRNA hypervariable tag datasets. Plos One 5(12):e15220. doi:10.1371/journal.pone.0015220
Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS (2009) Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75(16):5227–5236
Reeder J, Knight R (2009) The ‘rare biosphere’: a reality check. Nat Methods 6(9):636–637
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44(4):846–849
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405(6784):299–304
Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Postal G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin JY, Yen G, Schwartz DC, Welch RA, Blattner FR (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157: H7 (vol 409, pg 529, 2001). Nature 410(6825):240–240
Dembicki Jr. H, Samuels BM (2007) Identification, characterization, and groundtruthing of deepwater thermogenic hydrocarbon macroseepage utilizing high-resolution AUV geophysical data. In: Conference OT (ed) Offshore technology conference, Houston, TX, USA.
Dembicki Jr. H, Samuels BM (2008) Improving the detection and analysis of seafloor macro-seeps: an example from the Marco Polo Field, Gulf of Mexico, USA. In: International petroleum technology conference, Kuala Lumpur, Malaysia.
Ashby MN, Rine J, Mongodin EF, Nelson KE, Dimster-Denk D (2007) Serial analysis of rRNA genes and the unexpected dominance of rare members of microbial communities. Appl Environ Microbiol 73(14):4532–4542
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428(6978):37–43
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27(2):221–224. doi:10.1093/molbev/msp259
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, Russ C, Lander ES, Nusbaum C, Jaffe DB (2008) Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res 18(5):763–770
Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD (2011) Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77(11):3846–3852
Degnan PH, Ochman H (2012) Illumina-based analysis of microbial community diversity. ISME J 6(1):183–194
Acknowledgments
We thank Stewart Scherer for his bioinformatics assistance and helpful comments.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 101 kb)
Rights and permissions
About this article
Cite this article
Patin, N.V., Kunin, V., Lidström, U. et al. Effects of OTU Clustering and PCR Artifacts on Microbial Diversity Estimates. Microb Ecol 65, 709–719 (2013). https://doi.org/10.1007/s00248-012-0145-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0145-4