Abstract
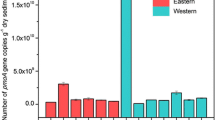
The distribution and phylogeny of extant protein-encoding genes recovered from geochemically diverse environments can provide insight into the physical and chemical parameters that led to the origin and which constrained the evolution of a functional process. Mercuric reductase (MerA) plays an integral role in mercury (Hg) biogeochemistry by catalyzing the transformation of Hg(II) to Hg(0). Putative merA sequences were amplified from DNA extracts of microbial communities associated with mats and sulfur precipitates from physicochemically diverse Hg-containing springs in Yellowstone National Park, Wyoming, using four PCR primer sets that were designed to capture the known diversity of merA. The recovery of novel and deeply rooted MerA lineages from these habitats supports previous evidence that indicates merA originated in a thermophilic environment. Generalized linear models indicate that the distribution of putative archaeal merA lineages was constrained by a combination of pH, dissolved organic carbon, dissolved total mercury and sulfide. The models failed to identify statistically well supported trends for the distribution of putative bacterial merA lineages as a function of these or other measured environmental variables, suggesting that these lineages were either influenced by environmental parameters not considered in the present study, or the bacterial primer sets were designed to target too broad of a class of genes which may have responded differently to environmental stimuli. The widespread occurrence of merA in the geothermal environments implies a prominent role for Hg detoxification in these environments. Moreover, the differences in the distribution of the merA genes amplified with the four merA primer sets suggests that the organisms putatively engaged in this activity have evolved to occupy different ecological niches within the geothermal gradient.



Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Anbar AD, Knoll AH (2002) Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297:1137–1142
Anbar AD, Duan Y, Lyons TW, Arnold GL, Kendall B, Creaser RA (2007) A wiff of oxygen before the great oxidation event? Science 317:1903–1906
Andersson A (1979) Mercury in soils. In: Nriagu O (ed) The biogeochemistry of mercury in the environment. Esevier, North Holland Biomedical Press, Amsterdam, pp 79–112
Ball JW, McCleskey RB, Nordstrom DK, Holloway JM (2007) Water-chemistry data for selected springs, geysers, and streams in Yellowstone National Park, Wyoming, 2003–2005. U.S. Geological Survey Open-File Report 2006–1339. U.S. Department of the Interior, City, pp. 183
Barkay T, Liebert C, Gillman M (1989) Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl Environ Microbiol 55:1574–1577
Barkay T, Gillman M, Turner RR (1997) Effects of dissolved organic carbon and speciation of Hg(II) on bioavailability of mercury. Appl Environ Microbiol 63:4267–4271
Barkay T, Kritee K, Boyd E, Geesey G (2010) A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ Microbiol 12:2904–2917
Boyd ES, Jackson RA, Encarnacion G, Zahn JA, Beard T, Leavitt WD, Pi Y, Zhang CL, Pearson A, Geesey GG (2007) Isolation, characterization, and ecology of sulfur-respiring crenarchaea inhabiting acid-sulfate-chloride-containing geothermal springs in Yellowstone National Park. Appl Environ Microbiol 73:6669–6677
Boyd ES, King S, Tomberlin JJ, Nordstrom DK, Krabbenhoft DP, Barkay T, Geesey GG (2009) Methylmercury enter an aquatic food web through acidophilic microbial mats in Yellowstone National Park, Wyoming. Environ Microbiol 11:950–959
Boyd ES, Hamilton TL, Spear JR, Lavin M, Peters JW (2010) [Fe-Fe]-hydrogenase in Yellowstone National Park: evidence for dispersal limitation and phylogenetic niche conservatism. ISME J 4:1485–1495
Bruce KD, Osborn AM, Pearson AJ, Strike P, Ritchie DA (1995) Genetic diversity within mer genes directly amplified from communities of noncultivated soil and sediment bacteria. Mol Ecol 4:605–612
Cai JH, Jia CQ (2010) Mercury removal from aqueous solutions using coke-derived sulfur-impregnated activated carbons. Ind Eng Chem Res 49:2716–2721
Crespo-Medina M, Chatziefthimiou AD, Bloom NS, Luther GW 3rd, Reinfelder JR, Vetriani C, Barkay T (2009) Adaptation of chemosynthetic microorganisms to elevated mercury concentrations in deep-sea hydrothermal vents. Limnol Oceanogr 54:41–49
Driscoll CT, Blette V, Yan C, Schofield CL, Munson R, Holsapple J (1995) The role of dissolved organic carbon in the chemistry and bioavailability of mercury in remote Adirondack lakes. Water Air Soil Pollut 80:499–508
Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345
Engle MA, Gustin MS, Goff F, Counce DA, Janik CJ, Bergfeld D, Rytuba JJ (2006) Atmospheric mercury emissions from substrates and fumaroles associated with three hydrothermal systems in the western United States. J Geophys Res-Atmosphere 111:D17304
Fietz S, Kobanova G, Izmest'eve L, Nicklisch A (2005) Regional, vertical, and seasonal distribution of phytoplankton and photosynthetic pigments in Lake Baikal. J Plankt Res 27:793–810
Hall BD, Olson ML, Rutter AP, Frontiera RR, Krabbenhoft DP, Gross DS, Yuen M, Rudolph TM, Schauer JJ (2006) Atmospheric mercury speciation in Yellowstone National Park. Sci Total Environ 367:354–366
Hamamura N, Olson SH, Ward DM, Inskeep WP (2005) Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl Environ Microbiol 71:5943–5950
Hirner AV, Krupp RE, Gainsford AR, Staerk H (1970) Metal-organic associations in sediments. II. Algal mats in contact with geothermal waters. Appl Geochem 5:507–513
Huber R, Sacher M, Vollmann A, Huber H, Rose D (2000) Respiration of arsenate and selenate by hyperthermophilic archaea. Syst Appl Microbiol 23:305–314
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Inskeep WP, McDermott TR (2007) Geomicrobiology of acid-sulfate-chloride springs in Yellowstone National Park. In: Inskeep WP, McDermott TR (eds) Geothermal biology and geochemistry in Yellowstone National Park. Montana State University, Bozeman, pp 143–162
Inskeep WP, Rusch DB, Jay Z, Herrgard MJ, Kozubal MA, Richardson TH, Macur RE, Hamamura N, Rd J, Fouke BW, Reysenbach A-L, Roberto F, Young M, Schwartz A, Boyd ES, Badger J, Mathur EJ, Ortmann AC, Bateson M, Geesey G, Frazier M (2010) Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773
King SA, Behnke S, Slack K, Krabbenhoft DP, Nordstrom DK, Burr MD, Striegl RG (2006) Mercury in water and biomass of microbial communities in hot springs of Yellowstone National Park, USA. Appl Geochem 21:1868–1879
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
McCleskey RB, Ball JW, Nordstrom DK, Holloway JM, Taylor HE (2004) Water-chemistry data for selected hot springs, geysers, and streams in Yellowstone National Park Wyoming 2001–2002. U.S. Geological Survey, U.S. Department of the Interior, Boulder, CO, 95 pages
Mergler D, Anderson HA, Chan LH, Mahaffey KR, Murray M, Sakamoto M, Stern AH (2007) Methylmercury exposure and health effects in humans: a worldwide concern. Ambio 36:3–11
Meyer-Dombard DR, Shock EL, Amend JP (2005) Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology 3:211–227
Morel FMM, Kraepiel AML (1998) The chemical cycle and bioaccumulation of mercury. Annu Rev Ecol Syst 29:543–566
Nabias JV, Carrott PJM, Carrott MMLR, Belchior M, Boavida D, Diall T, Gulyurtlu I (2006) Mercury removal from aqueous solution and flue gas by adsorption on activated carbon fibres. Appl Surf Sci 252:6046–6052
Nakagawa R (1974) The mercury content in hot springs. Nippon Kagaku Kaishi:71–74
Nakagawa R (1984) Amounts of mercury discharged to atmosphere from fumarols and hot spring gases in geothermal areas. Nippon Kagaku Kaishi:709–715
Nakagawa R (1999) Estimation of mercury emission from geothermal activity in Japan. Chemosphere 38:1867–1871
Nazaret S, Jeffrey WH, Saouter E, Von Haven R, Barkay T (1994) merA gene expression in aquatic environments measured by mRNA production and Hg(II) volatilization. Appl Environ Microbiol 60:4059–4065
Newton RJ, Jones SE, Helmus MR, McMahon KD (2007) Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol 73:7169–7176
Ní Chadhain SM, Schaefer JK, Crane S, Zylstra GJ, Barkay T (2006) Analysis of mercuric reductase (merA) gene diversity in an anaerobic mercury-contaminated sediment enrichment. Environ Microbiol 8:1746–1752
Nies DH (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339
Nriagu J, Becker C (2003) Volcanic emissions of mercury to the atmosphere: global and regional inventories. Sci Total Environ 304:3–12
Nriagu JO (1989) A global assessment of natural sources of atmospheric trace metals. Nature 338:47–49
Nunoura T, Hirayama H, Takami H, Oida H, Nishi S, Shimamura S, Suzuki Y, Inagaki F, Takai K, Nealson KH, Horikoshi K (2005) Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ Microbiol 7:1967–1984
Øregaard G, Sørensen SJ (2007) High diversity of bacterial mercuric reductase genes from surface and sub-surface floodplain soil (Oak Ridge, USA). ISME J 1:453–467
Palsson B (2011) Adaptive laboratory evolution. Microbe 6:69–74
Phelps D, Buseck PR (1980) Distribution of soil mercury and the development of soil mercury anomalies in the Yellowstone geothermal area, Wyoming. Econ Geol 75:730–741
Poulain AJ, Ní Chadhain SM, Ariya PA, Amyot M, Garcia E, Campbell PGC, Zylstra GJ, Barkay T (2007) A potential for mercury reduction by microbes in the High Arctic. Appl Environ Microbiol 73:2230–2238
Ramus J, Beale SI, Mauzerall D, Howard KL (1976) Changes in photosynthetic pigment concentration in seaweeds as a function of water depth. Mar Biol 37:223–229
Ravichandran M (2004) Interactions between mercury and dissolved organic matter—a review. Chemosphere 55:319–331
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rooney JP (2007) The role of thiols, dithiols, nutritional factors and interacting ligands in the toxicity of mercury. Toxicology 234:145–156
Sabehi G, Kirkup BC, Rozenberg M, Stambler N, Polz MF, Beja O (2007) Adaptation of spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and Sargasso Seas. ISME J 1:48–55
Saito MA, Sigman DM, Morel FMM (2003) The bioinorganic chemistry of the ancient ocean: the co-evolution of the cyanobacterial metal requirements and biogeochemical cycles in the Archaean-Proterozoic boundary? Inorg Chim Acta 356:308–318
Schaefer JK, Yagi J, Reinfelder JR, Cardona T, Ellickson KM, Tel-Or S, Barkay T (2004) Role of the bacterial organomercury lyase (MerB) in controlling methylmercury accumulation in mercury-contaminated natural waters. Environ Sci Technol 38:4304–4311
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schuster E (1991) The behavior of mercury in the soil with special emphasis on comlexation and adsorption proceses—a review of the literature. Water Air Soil Pollut 56:667–680
Sherman LS, Blum JD, Nordstrom DK, McCleskey RB, Barkay T, Vetriani C (2009) Mercury isotopic composition of hydrothermal systems in the Yellowstone Plateau volcanic field and Guaymas Basin sea-floor rift. Earth Planet Sci Lett 279:86–96
Siciliano SD, O'Driscoll NJ, Lean DR (2002) Microbial reduction and oxidation of mercury in freshwater lakes. Environ Sci Technol 36:3064–3068
Simbahan J, Kurth E, Schelert J, Dillman A, Moriyama E, Jovanovich S, Blum P (2005) Community analysis of a mercury hot spring supports occurrence of domain-specific forms of mercuric reductase. Appl Environ Microbiol 71:8836–8845
Spear JR, Walker JJ, McCollom TM, Pace NR (2005) Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc Nat Acad Sci USA 102:2555–2560
Stoffers P, Hannington M, Wright I, Herzig P, de Ronde E (1999) Elemental mercury at submarine hydrothermal vents in the Bay of Plenty, Taupo volcanic zone, New Zealand. Geology 27:931–934
Swofford DL (2003) PAUP*: Phylogenetic analysis using parsimony version 4.0b10 (*and other methods). Sinauer Associates, Sunderland
Team RDC (2010) R Foundation for Statistical Computing. Austria, Vienna
Vetriani C, Chew YS, Miller SM, Yagi J, Coombs J, Lutz RA, Barkay T (2005) Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl Environ Microbiol 71:220–226
Yamamoto M (1996) Stimulation of elemental mercury oxidation in the presence of chloride ion in aquatic environments. Chemosphere 32:1217–1224
Zillig W, Stetter KO, Schafer W, Janekovic D, Wunderl S, Holz I, Palm P (1981) Thermoproteales: a novel type of extremely thermoacidophilic anaerobic archaebacteria isolated from Icelandic solfataras. Zentralbl Mikrobiol Parasitenkd Infektionskr Hyg Abt 1 Orig C2:205–227
Acknowledgments
We thank Jesse Bennett and Kim Slack for assisting with fieldwork in YNP, John DeWild and personnel of USGS mercury Lab (Middleton, WI) for supporting Hg analyses, and Christie Hendrix and her colleagues at the YNP Research Permit Office for their enthusiastic support and field access. This research was supported by the Thermal Biology Institute at Montana State University, through NASA award NAG5-8807: Center for Studying Life in Extreme Environments, by the Environmental Remediation Science Program (ERSP), Biological and Environmental Research (BER), U.S. Department of Energy (Grant DE-FG02-05ER63969), by a European Union Marie Curie Actions—International Incoming Fellowship (FP7-PEOPLE-IIF-2008), and by the National Science Foundation Microbial Observatories program (grant MCB0132022). ESB was supported by an Inland Northwest Research Alliance Graduate Fellowship grant DE-FG07-02ID14277, a Montana University System Water Center Fellowship, and a NASA Astrobiology Institute postdoctoral fellowship. ESB also acknowledges support for the Astrobiology Biogeocatalysis Research Center from the NAI.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Table S1
Primer sets and respective PCR conditions used to target diversity of merA sequences in samples used for this study (DOC 38 kb)
Table S2
Specificity range of the six degenerate merA primer sets (DOC 42 kb)
Table S3
Clones constructed from amplicons generated with primer sets 2, 3, and 4 from Hg-containing sites in geothermal springs of Yellowstone National Park (XLS 38 kb)
Figure S1
A neighbor-joining tree of MerA sequences used for the design of the merA primer sets. MerA protein sequences were downloaded from the NCBI database. The sequences were aligned using ClustalW2.0, trimmed in ClustalX, and a the neighbor-joining phylogenetic tree were constructed using PAUP* 4.0 (Swofford, D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony [*and Other Methods]. Version 4. Sinauer Associates, Sunderland, Massachusetts) with 1, 000 bootstrap replicates. The range of organisms whose merA genes were targeted by each primer set is indicated by brackets. Numbers alongside brackets correspond to the PCR primer set designed to amplify merA sequences in the bracketed species. The asterisk following A. pernix K1 denotes the merA of this organism was a part of the alignments that were used for the design of both primer sets 5 and 6. Bar—50 base substitutions. Bootstrap values are indicated at branching points. Underlined species name implies that mercuric reductase activity was documented in this species. (DOCX 76 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Boyd, E., Crane, S. et al. Environmental Conditions Constrain the Distribution and Diversity of Archaeal merA in Yellowstone National Park, Wyoming, U.S.A.. Microb Ecol 62, 739–752 (2011). https://doi.org/10.1007/s00248-011-9890-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9890-z