Abstract
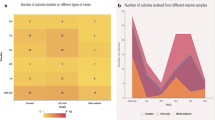
Marine environment habitats, such as the coral mucus layer, are abundant in nutrients and rich with diverse populations of microorganisms. Since interactions among microorganisms found in coral mucus can be either mutualistic or competitive, understanding quorum sensing-based acyl homoserine lactone (AHL) language may shed light on the interaction between coral-associated microbial communities in the native host. More than 100 bacterial isolates obtained from different coral species were screened for their ability to produce AHL. When screening the isolated coral bacteria for AHL induction activity using the reporter strains Escherichia coli K802NR-pSB1075 and Agrobacterium tumefaciens KYC55, we found that approximately 30% of the isolates tested positive. Thin layer chromatography separation of supernatant extracts revealed different AHL profiles, with detection of at least one active compound in the supernatant of those bacterial extracts being able to induce AHL activity in the two different bioreporter strains. The active extract of bacterial isolate 3AT 1-10-4 was subjected to further analysis by preparative thin layer chromatography and liquid chromatography tandem mass spectrometry. One of the compounds was found to correspond with N-(3-hydroxydecanoyl)-l-homoserine lactone. 16S rRNA gene sequencing of the isolates with positive AHL activity affiliated them with the Vibrio genus. Understanding the ecological role of AHL in the coral environment and its regulatory circuits in the coral holobiont-associated microbial community will further expand our knowledge of such interactions.





Similar content being viewed by others
References
Atkinson S, Williams P (2009) Quorum sensing and social networking in the microbial world. J R Soc Interface 6:959–978
Bourne D, Iida Y, Uthicke S, Smith-Keune C (2008) Changes in coral-associated microbial communities during a bleaching event. ISME J 2:350–363
Bruhn JB, Nielsen KF, Hjelm M, Hansen M, Bresciani J, Schulz S, Gram L (2005) Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl Environ Microbiol 71:7263–7270
Camilli A, Bassler BL (2006) Bacterial small-molecule signaling pathways. Science 311:1113–1116
Decho AW, Norman RS, Visscher PT (2010) Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol 18:73–80
Dubern JF, Diggle SP (2008) Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst 4:882–888
Dunne WM Jr (2002) Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15:155–166
Eberhard A, Widrig CA, McBath P, Schineller JB (1986) Analogs of the autoinducer of bioluminescence in Vibrio fischeri. Arch Microbiol 146:35–40
Friedrich AB, Merkert H, Fendert T, Hacker J, Proksch P, Hentschel U (1999) Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridization (FISH). Marine Biology 134:461–470
Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468
Glick R, Gilmour C, Tremblay J, Satanower S, Avidan O, Deziel E, Greenberg EP, Poole K, Banin E (2010) Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 192:2973–2980
Hakkila K, Maksimow M, Karp M, Virta M (2002) Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal Biochem 301:235–242
Harel M, Ben-Dov E, Rasoulouniriana D, Siboni N, Kramarsky-Winter E, Loya Y, Ze B, Wiesman Z, Kushmaro A (2008) A new Thraustochytrid, strain Fng1, isolated from the surface mucus of the hermatypic coral Fungia granulosa. FEMS Microbiol Ecol 64:378–387
Huang YL, Ki JS, Lee OO, Qian PY (2009) Evidence for the dynamics of Acyl homoserine lactone and AHL-producing bacteria during subtidal biofilm formation. ISME J 3:296–304
Jayaraman A, Wood TK (2008) Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng 10:145–167
Jensen PR, Kauffman CA, Fenical W (1996) High recovery of culturable bacteria from the surfaces of marine algae. Marine Biology 126:1–7
Joint I, Tait K, Wheeler G (2007) Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos Trans R Soc Lond B Biol Sci 362:1223–1233
Kooperman N, Ben-Dov E, Kramarsky-Winter E, Barak Z, Kushmaro A (2007) Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol Lett 276:106–113
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kumari A, Pasini P, Daunert S (2008) Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal Bioanal Chem 391:1619–1627
Kuo A, Callahan SM, Dunlap PV (1996) Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol 178:971–976
Kushmaro A, Loya Y, Fine M, Rosenberg E (1996) Bacterial infection and coral bleaching. Nature 380:396–396
Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4975–4983
Lyon GJ, Novick RP (2004) Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25:1389–1403
Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S (1999) Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145(Pt 2):283–291
Middleton B, Rodgers HC, Camara M, Knox AJ, Williams P, Hardman A (2002) Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol Lett 207:1–7
Milton DL (2006) Quorum sensing in vibrios: complexity for diversification. Int J Med Microbiol 296:61–71
Mohamed NM, Cicirelli EM, Kan J, Chen F, Fuqua C, Hill RT (2008) Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol 10:75–86
Ng W-L, Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222
Passador L, Tucker KD, Guertin KR, Journet MP, Kende AS, Iglewski BH (1996) Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol 178:5995–6000
Patriquin GM, Banin E, Gilmour C, Tuchman R, Greenberg EP, Poole K (2008) Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 190:662–671
Pesci EC, Pearson JP, Seed PC, Iglewski BH (1997) Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 179:3127–3132
Rampioni G, Bertani I, Zennaro E, Polticelli F, Venturi V, Leoni L (2006) The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter. J Bacteriol 188:815–819
Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E (2006) The coral probiotic hypothesis. Environ Microbiol 8:2068–2073
Ritchie BK (2006) Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14
Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N (2001) Diversity of bacteria associated with the Caribbean coral Montastraeafranksi. Coral Reefs 20:85–91
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sharon G, Rosenberg E (2008) Bacterial growth on coral mucus. Curr Microbiol 56:481–488
Shnit-Orland M, Kushmaro A (2009) Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol 67:371–380
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tait K, Hutchison Z, Thompson FL, Munn CB (2010) Quorum sensing signal production and inhibition by coral-associated vibrios. Environ Microbiol Rep 2:145–150
Taylor MW, Schupp PJ, Baillie HJ, Charlton TS, de Nys R, Kjelleberg S, Steinberg PD (2004) Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl Environ Microbiol 70:4387–4389
Thenmozhi R, Nithyanand P, Rathna J, Pandian SK (2009) Antibiofilm activity of coral-associated bacteria against different clinical M serotypes of Streptococcus pyogenes. FEMS Immunol Med Microbiol 57:284–294
Wahl M (1995) Bacterial epibiosis on Bahamian and Pacific ascidians. J Exp Mar Biol Ecol 191:239–255
Webster NS, Wilson KJ, Blackall LL, Hill RT (2001) Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl Environ Microbiol 67:434–444
Williams P (2007) Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153:3923–3938
Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GS (1998) Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett 163:185–192
Zhu J, Chai Y, Zhong Z, Li S, Winans SC (2003) Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol 69:6949–6953
Acknowledgments
This work was supported by the National Institute of Biotechnology in the Negev (NIBN), with financial support under grant number 8528620 “Bioactive compounds”, ISF Grant 1169/07, and Marie Curie Host Fellowships for the Transfer of Knowledge (TOK) Development Host Scheme (MEMBIOF) contract no. MTKD-CT-2005-029813. The authors also thank the IUI, Eilat, Israel, for use of their facilities, Prof Jun Zhu for strain KYC55, and Dr. Tamar Amir, Dr. Eitan Ben-Dov, Dr. Michal Shani Sekler, Luba Arotsker, and Nahshon Siboni for sample collection, technical support, and guidance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Golberg, K., Eltzov, E., Shnit-Orland, M. et al. Characterization of Quorum Sensing Signals in Coral-Associated Bacteria. Microb Ecol 61, 783–792 (2011). https://doi.org/10.1007/s00248-011-9848-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9848-1