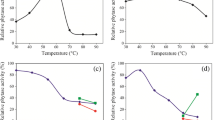
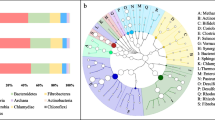
Abstract
Examples of a new class of phytase related to protein tyrosine phosphatases (PTP) were recently isolated from several anaerobic bacteria from the rumen of cattle. In this study, the diversity of PTP-like phytase gene sequences in the rumen was surveyed by using the polymerase chain reaction (PCR). Two sets of degenerate primers were used to amplify sequences from rumen fluid total community DNA and genomic DNA from nine bacterial isolates. Four novel PTP-like phytase sequences were retrieved from rumen fluid, whereas all nine of the anaerobic bacterial isolates investigated in this work contained PTP-like phytase sequences. One isolate, Selenomonas lacticifex, contained two distinct PTP-like phytase sequences, suggesting that multiple phytate hydrolyzing enzymes are present in this bacterium. The degenerate primer and PCR conditions described here, as well as novel sequences obtained in this study, will provide a valuable resource for future studies on this new class of phytase. The observed diversity of microbial phytases in the rumen may account for the ability of ruminants to derive a significant proportion of their phosphorus requirements from phytate.



Similar content being viewed by others
References
Altschul, SF, Gish, W, Miller, W, Myers, EW, Lipman, DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410
Barford, D, Flint, AJ, Tonks, NK (1994) Crystal structure of human protein tyrosine phosphatase 1B. Science 263: 1397–1404
Bravo, D, Meschy, F, Bogaert, C, Sauvant, D (2002) Effects of fungal phytase addition, formaldehyde treatment and dietary concentrate content on ruminal phosphorus availability. Anim Feed Sci Technol 99: 73–95
Bravo, D, Sauvant, D, Bogaert, C, Meschy, F (2003) II. Quantitative aspects of phosphorus absorption in ruminants. Reprod Nutr Dev 43: 271–284
Bryant, MP, Burkey, LA (1953) Cultural methods and some characteristics of some of the numerous groups of bacteria in the bovine rumen. J Dairy Sci 36: 205–217
Chu, H-M, Guo, R-T, Lin, T-W, Chou, C-C, Shr, H-L, Lai, H-L, Tang, T-Y, Cheng, K-J, Selinger, BL, Wang, AHJ (2004) Structures of Selenomonas ruminantium phytase in complex with persulfated phytate: DSP phytase fold and mechanism for sequential substrate hydrolysis. Structure 12: 2015
Cosgrove, DJ (1966) The chemistry and biochemistry of inositol polyphosphates. Rev Pure Appl Chem 16: 209–215
Graf, E (1986) Chemistry and applications of phytic acid: an overview. In: Graf, E (Ed.) Phytic Acid, Chemistry and Applications. Pilatus Press, Minneapolis, pp 1–22
Heinkoff, S, Heinkoff, JG, Alfrod, WJ, Pitrokovski, S (1995) Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163: 12–26
Higgins, D, Thompson, J, Gibson, T, Thompson, JD (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680
Hungate, RE (1950) The anaerobic mesophyllic cellulolytic bacteria. Bacteriol Rev 14: 1–49
Konietzny, U, Greiner, R (2002) Molecular and catalytic properties of phytate-degrading enzymes (phytases). Int J Food Sci Technol 37: 791–812
McAllister, TA, Forster, RJ, Teather, RM, Sharma, R, Attwood, GT, Selinger, LB, Joblin, KN (2005) Manipulation and characterization of the rumen ecosystem through biotechnology. In: Mosenthin, R, Zentek, J, Zebrowska, T (Eds.) Biology of Nutrition in Growing Animals, vol. 4, Elsevier, Amsterdam, pp 559–583
Morse, D, Head, HH, Wilcox, CJ (1992) Disappearance of phosphorus in phytate from concentrates in vitro and from rations fed to lactating dairy cows. J Dairy Sci 75: 1979–1986
Mullaney, EJ, Daly, CB, Ullah, AHJ (2000) Advances in phytase research. Adv Appl Microbiol 47: 157–199
Mullaney, EJ, Ullah, AHJ (2003) The term phytase comprises several different classes of enzymes. Biochem Biophys Res Commun 312: 179–184
Nayini, NR, Markakis, P (1986) Phytases. In: Graf, E (Ed.) Phytic Acid, Chemistry and Applications. Pilatus Press, Minneapolis, pp 101–118
Nelson, TS, Daniels, LB, Hall, JR, Shields, LG (1976) Hydrolysis of natural phytate phosphorus in the digestive tract of calves. J Anim Sci 42: 1509–1512
Nicholas, KB, Nicholas, HB, Deerfield, DW. GeneDoc: analysis and visualization of genetic variation. Embnet News 4 : 14
Polz, MF, Cavanaugh, CM (1998) Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64: 3724–3730
Priefer, U, Simon, R, Puhler, A (1984) Cloning with cosmids. In: Puhler, A, Timmis, KN (Eds.) Advanced Molecular Genetics. Springer-Verlag, New York, pp 190–201
Raun, A, Cheng, E, Burroughs, W (1956) Phytate phosphorus hydrolysis and availability to rumen microorganisms. Agric Food Chem 4: 869–871
Rose, TM, Schultz, ER, Heinkoff, JG, Pietrokovski, S, McCallum, CM, Heinkoff, S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26: 1628–1635
Scott, HW, Dehority, BA (1965) Vitamin requirements of several cellulolytic bacteria. J Bacteriol 89: 1169–1175
Selinger, LB, Forsberg, CW, Cheng, KJ (1996) The rumen: a unique source of enzymes for enhancing livestock production. Anaerobe 2: 263–284
Swofford, DL (2003) PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) (4.0 ed.). Sinauer Associates, Sunderland, MA
Yanke, LJ, Bae, HD, Selinger, LB, Cheng, K-J (1998) Phytase activity of anaerobic ruminal bacteria. Microbiology 144: 1565–1573
Acknowledgments
The authors thank Jay Yanke, Bob Williams and Bob Forster. An NSERC operating grant to L.B.S. and an NSERC undergraduate student research award to B.A.N. supported this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nakashima, B.A., McAllister, T.A., Sharma, R. et al. Diversity of Phytases in the Rumen. Microb Ecol 53, 82–88 (2007). https://doi.org/10.1007/s00248-006-9147-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9147-4