Abstract
Background
A radiopaque marker study measures colonic transit time for work-up of primary constipation. It requires the patient to ingest multiple tiny radiopaque markers, which the radiologist must count manually on follow-up abdominal radiographs. Counting these markers is tedious but cognitively simple.
Objective
To develop a convolutional neural network (CNN) capable of counting the number of radiopaque markers on abdominal radiographs.
Materials and methods
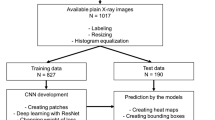
The image archive at a large tertiary children’s hospital was searched to identify abdominal radiographs performed in children for the indication of a radiopaque marker study. To establish the ground truth, a radiologist manually labeled the coordinates of the radiopaque markers in each radiograph and thereby generated a density map for that radiograph. A CNN was trained to estimate this density map from its corresponding abdominal radiograph. Spatially integrating the output density map provided an estimate of the number of markers in the radiograph. To assess model accuracy, mean absolute error and root mean square error were calculated.
Results
The study cohort consisted of 436 radiographs (mean number of markers per radiograph: 34). This cohort was randomly divided into training, validation and testing sets consisting of 306, 65 and 65 radiographs, respectively. Based on the testing set, the CNN accurately estimated the number of markers in each radiograph with mean absolute error=2.6 markers and root mean square error=3.9 markers.
Conclusion
The proposed CNN generated promising results in counting the number of radiopaque markers on abdominal radiographs and offers the potential of automating the interpretation of colonic transit studies.







Similar content being viewed by others
References
van den Berg MM, Benninga MA, Di Lorenzo C (2006) Epidemiology of childhood constipation: a systemic review. Am J Gastroenterol 101:2401–2409
Robin SG, Keller C, Zwiener R et al (2018) Prevalence of pediatric functional gastrointestinal disorders utilizing the Rome IV criteria. J Pediatr 195:134–139
Andrews CN, Storr M (2011) The pathophysiology of chronic constipation. Can J Gastroenterol 25(Suppl B):16B–21B
Tabbers MM, Dilorenzo C, Berger MY et al (2014) Evaluation and treatment of functional constipation in infants and children: evidence-based recommendations from ESPGHAN and NASPGHAN. J Pediatr Gastroenterol Nutr 58:258–274
Locke GR 3rd, Pemberton JH, Phillips SF (2000) AGA technical review on constipation. American Gastroenterological Association. Gastroenterology 119:1766–1778
Hinton JM, Lennard-Jones JE, Young AC (1969) A new method for studying gut transit times using radiopaque markers. Gut 10:842–847
Arhan P, Devroede G, Jehannin B et al (1981) Segmental colonic transit time. Dis Colon Rectum 24:625–629
Corazziari E, Cucchiara S, Staiano A et al (1985) Gastrointestinal transit time, frequency of defecation, and anorectal manometry in healthy and constipated children. J Pediatr 106:379–382
Bautista Casasnovas A, Varela Cives R, Villanueva Jeremias A et al (1991) Measurement of colonic transit time in children. J Pediatr Gastroenterol Nutr 13:42–45
Zaslavsky C, da Silveira TR, Maguilnik I (1998) Total and segmental colonic transit time with radio-opaque markers in adolescents with functional constipation. J Pediatr Gastroenterol Nutr 27:138–142
Gutierrez C, Marco A, Nogales A, Tebar R (2002) Total and segmental colonic transit time and anorectal manometry in children with chronic idiopathic constipation. J Pediatr Gastroenterol Nutr 35:31–38
Wagener S, Shankar KR, Turnock RR et al (2004) Colonic transit time—what is normal? J Pediatr Surg 39:166–169
Xie W, Noble JA, Zisserman A (2018) Microscopy cell counting and detection with fully convolutional regression networks. Comp Methods Biomech Biomed Eng: Imaging Visual 6:283–292
Tessema AW, Mohammed MA, Simegn GL, Kwa TC (2021) Quantitative analysis of blood cells from microscopic images using convolutional neural network. Med Biol Eng Comput 59:143–152
Yuzkat M, Ilhan HO, Aydin N (2021) Multi-model CNN fusion for sperm morphology analysis. Comput Biol Med 137:104790
Ilyas N, Shahzad A, Kim K (2019) Convolutional-neural network-based image crowd counting: review, categorization, analysis, and performance evaluation. Sensors (Basel) 20:43
Afonso M, Fonteijn H, Fiorentin FS et al (2020) Tomato fruit detection and counting in greenhouses using deep learning. Front Plant Sci 11:571299
Padubidri C, Kamilaris A, Karatsiolis S, Kamminga J (2021) Counting sea lions and elephants from aerial photography using deep learning with density maps. Animal Biotelemetry 9:9–27
Metcalf AM, Phillips SF, Zinsmeister AR et al (1987) Simplified assessment of segmental colonic transit. Gastroenterology 92:40–47
American College of Radiology (2021) ACR-SAR-SPR practice parameter for the performance of abdominal radiograph (Res. 39). In: American College of Radiology. ACR Standards. Reston VA: American College of Radiology
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, pp 234–241
Mantelli H, Devroede G, Arhan P, Duguay E (1978) Mechanisms of idiopathic constipation: outlet obstruction. Gastroenterology 75:623–631
Waite A, Devroede G, Duranceau A et al (1983) Constipation with colonic inertia: a manifestation of systemic disease? Dig Dis Sci 28:1025–1033
Arhan P, Devroede G, Jehannu B et al (1981) Segmental colonic transit time. Dis Colon Rectum 24:625–629
Chaussade S, Roche H, Khyari A et al (1986) Mesure du temps de transit colique (TTC): description et validation d’une nouvelle technique [Measurement of colonic transit time: description and validation of a new method]. Gastroenterol Clin Biol 10:385–389
Casasnovas AB, Cives RV, Jeremias AV et al (1991) Measurement of colonic transit time in children. J Pediatr Gastroenterol Nutr 13:42–45
Pan SJ, Yang Q (2018) A survey on transfer leaning. IEEE Trans Knowl Data Eng 22:1345–1359
Refaeilzadeh P, Tang L, Liu H (2009) Cross-validation. In: Liu L, Özsu MT (eds) Encyclopedia of database systems. Springer, Boston
Acknowledgements
The author would like to thank Ms. Nancy Drinan for her help in the editing of the manuscript. The author would also like to acknowledge the use of Boston Children’s Hospital’s High-Performance Computing Resources BCH HPC Cluster Enkefalos 2 (E2) which has been crucial to the research reported in this publication. Software used in the project was installed and configured by BioGrids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tsai, A. Density map estimation with convolutional neural networks to count radiopaque markers on colonic transit studies. Pediatr Radiol 52, 2178–2187 (2022). https://doi.org/10.1007/s00247-022-05371-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00247-022-05371-1