Abstract
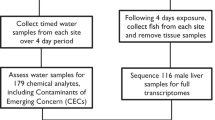
It is postulated that below a transcriptomic-based point of departure, adverse effects are unlikely to occur, thereby providing a chemical concentration to use in screening level hazard assessment. The present study extends previous work describing a high-throughput fathead minnow assay that can provide full transcriptomic data after exposure to a test chemical. One-day post-hatch fathead minnows were exposed to ten concentrations of three representatives of four chemical modes of action: organophosphates, ecdysone receptor agonists, plant photosystem II inhibitors, and estrogen receptor agonists for 24 h. Concentration response modeling was performed on whole body gene expression data from each exposure, using measured chemical concentrations when available. Transcriptomic points of departure in larval fathead minnow were lower than apical effect concentrations across fish species but not always lower than toxic effect concentrations in other aquatic taxa like crustaceans and insects. The point of departure was highly dependent on measured chemical concentration which were often lower than the nominal concentration. Differentially expressed genes between chemicals within modes of action were compared and often showed statistically significant overlap. In addition, reproducibility between identical exposures using a positive control chemical (CuSO4) and variability associated with the transcriptomic point of departure using in silico sampling were considered. Results extend a transcriptomic-compatible fathead minnow high-throughput assay for possible use in ecological hazard screening.



Similar content being viewed by others
Data Availability
All sequence data reported in this manuscript are accessible via the Gene Expression Omnibus (BioProject PRJNA104613).
References
Ankley GT, Johnson RD (2004) Small fish models for identifying and assessing the effects of endocrine-disrupting chemicals. ILAR J 45(4):469–483. https://doi.org/10.1093/ilar.45.4.469
Ankley GT, Villeneuve DL (2006) The fathead minnow in aquatic toxicology: past, present and future. Aquat Toxicol 78(1):91–102. https://doi.org/10.1016/j.aquatox.2006.01.018
Ankley GT, Jensen KM, Kahl MD, Makynen EA, Blake LS, Greene KJ, Johnson RD, Villeneuve DL (2007) Ketoconazole in the fathead minnow (Pimephales promelas): reproductive toxicity and biological compensation. Environ Toxicol Chem 26(6):1214–1223. https://doi.org/10.1897/06-428R.1
Ankley GT, LaLone CA, Gray LE, Villeneuve DL, Hornung MW (2016) Evaluation of the scientific underpinnings for identifying estrogenic chemicals in nonmammalian taxa using mammalian test systems. Environ Toxicol Chem 35(11):2806–2816. https://doi.org/10.1002/etc.3456
Ankley GT, Feifarek D, Blackwell BR, Cavallin JE, Jensen KM, Kahl MD, Poole S, Randolph E, Saari T, Villeneuve DL (2017) Re-evaluating the significance of estrone as an environmental estrogen. Environ Sci Tech 51(8):4705–4713. https://doi.org/10.1021/acs.est.7b00606
Bhandari RK, Deem S, Holliday D, Jandegian C, Kassotis C, Nagel S, Tillitt DE, vom Saail FS, Rosenfeld CS (2015) Effects of the environmental estrogenic contaminants bisphenol A and 17α-ethinyl estradiol on sexual development and adult behaviors in aquatic wildlife species. Gen Comp Endocrin 214:195–219. https://doi.org/10.1016/j.ygcen.2014.09.014
Chagas AP, Peixoto BP, da Costa BB, Moreira TA, Cinelli LP, da Silva LL, Miranda-Alves L, Berto-Junior C (2021) Effects of bisphenol A and S on blood coagulation: in vivo, in vitro and in silico approaches in toxicodynamic. Toxicol Mech Methods 31(2):90–99. https://doi.org/10.1080/15376516.2020.1836102
Crago J, Corsi SR, Weber D, Bannerman R, Klaper R (2011) Linking biomarkers to reproductive success of caged fathead minnows in streams with increasing urbanization. Chemosphere 82(11):1669–1674. https://doi.org/10.1016/j.chemosphere.2010.11.011
Dean M, Annilo T (2005) Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 6:123–142. https://doi.org/10.1146/annurev.genom.6.080604.162122
EURL (2021) EURL ECVAM activies. Alt Lab Animals 49(6):229–234. https://doi.org/10.1177/02611929211063560.jrc
Feswick A, Isaacs M, Biales A, Flick RW, Bencic DC, Wang RL, Vulpe C, Brown-Augustine M, Loguinov A, Falciani F, Antczak P, Herbert J, Brown L, Denslow ND, Kroll KJ, Lavelle C, Dang V, Escalon L, Garcia-Reyero N, Martyniuk CJ, Munkittrick KR (2017) How consistent are we? Interlaboratory comparison study in fathead minnows using the model estrogen 17α-ethinylestradiol to develop recommendations for environmental transcriptomics. Environ Toxicol Chem 36(10):2614–2623. https://doi.org/10.1002/etc.3799
Hao R, Bondesson M, Singh AV, Riu A, McCollum CW, Knudsen TB, Gorelick DA, Gustafsson JA (2013) Identification of estrogen target genes during zebrafish embryonic development through transcriptomic analysis. PLoS ONE 8(11):e79020. https://doi.org/10.1371/journal.pone.0079020
Harrill JA, Everett LJ, Haggard DE, Sheffield T, Bundy JL, Willis CM, Thomas RS, Shah I, Judson RS (2021) High-throughput transcriptomics platform for screening environmental chemicals. Toxicol Sci 181(1):68–89. https://doi.org/10.1093/toxsci/kfab009
Horimizu R, Ogawa R, Watanabe Y, Tatsukawa H, Kinoshita M, Hashimoto H, Hitomi K (2017) Biochemical characterization of a medaka (Oryzias latipes) orthologue for mammalian factor XIII and establishment of a gene-edited mutant. The FEBS J 284(17):2843–2855. https://doi.org/10.1111/febs.14153
Huff M, da Silveira W, Hazard ES, Courtney SM, Rednaud L, Hardiman G (2019) Systems analysis of the liver transcriptome in adult male zebrafish exposed to the non-ionic surfactant nonylphenol. Gen Comp Endocrinol 271:1–14. https://doi.org/10.1016/j.ygcen.2018.10.016
Johnson KJ, Auerbach SS, Stevens T, Barton-Maclaren TS, Costa E, Currie RA, Wilk DD, Haq S, Rager JE, Reardon AJF, Wehmas L, Williams A, O’Brien J, Yauk C, LaRocca JL, Pettit S (2022) A transformative vision for an omics-based regulatory chemical testing paradigm. Toxicol Sci 190(2):127–132. https://doi.org/10.1093/toxsci/kfac097
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acid Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Kavlock RJ, Chandler K, Houck KA, Hunter S, Judson RS, Kleinstreuer NC, Knudsen T, Martin M, Padilla S, Reif D, Richard A, Rotroff D, Sipes N, Dix D (2012) Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25(7):1287–1302. https://doi.org/10.1021/tx3000939
Kavlock RJ, Daston GP, DeRosa C, Fenner-Crisp PA, Gray LE, Kaattari SL, Lucier G, Luster M, Mac MJ, Maczka C, Miller R, Moore J, Rolland R, Scott G, Sheehan DM, Tilson HA (1996) Research needs for the risk assessment of health and environmental effects of endocrine disruptors: a report of the U.S. EPA-sponsored workshop. Environ Health Perspect 104(4): 715–740. https://doi.org/10.1289/ehp.96104s4715
Knigge T, LeBlanc GA, Ford AT (2021) A crab Is not a fish: unique aspects of the crustacean endocrine system and considerations for endocrine toxicology. Front Endocrinol 12. https://doi.org/10.3389/fendo.2021.587608
Krewski D, Acosta D Jr, Andersen M, Anderson H, Bailer JC III, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S Jr, Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L (2010) Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Environ Health B Crit Rev 13(2–4):51–138. https://doi.org/10.1080/10937404.2010.483176
Loughery JR, Marentette JR, Frank RA, Hewitt M, Parrott JL, Martyniuk CJ (2019) Transcriptome profiling in larval fathead minnow exposed to commercial naphthenic acids and extracts from fresh and aged oil sands process-affected water. Environ Sci Technol 53(17):10435–10444. https://doi.org/10.1021/acs.est.9b01493
Love M, Anders S, Huber W (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(550). https://doi.org/10.1186/s13059-014-0550-8
Luijten M, Wackers PFK, Rorije E, Pennings JLA, Heusinkveld HJ (2021) Relevance of in vitro transcriptomics for in vivo mode of action assessment. Chem Res Toxicol 34(2):452–459. https://doi.org/10.1021/acs.chemrestox.0c00313
Martinson JW, Bencic DC, Toth GP, Kostich MS, Flick RW, See MJ, Lattier D, Biales AD, Huang W (2022) De novo assembly of the nearly complete fathead minnow reference genome reveals a repetitive but compact genome. Environ Toxicol Chem 41(2):448–461. https://doi.org/10.1002/etc.5266
Mittal K, Ewald J, Basu N (2022) Transcriptomic points of departure calculated from rainbow trout gill, liver, and gut cell lines exposed to methylmercury and fluoxetine. Environ Toxicol Chem 41(8):1982–1992. https://doi.org/10.1002/etc.5395
Morgenstern R, Zhang J, Johansson K (2011) Microsomal glutathione transferase 1: mechanism and functional roles. Drug Metabol Rev 43(2):300–306. https://doi.org/10.3109/03602532.2011.558511
NRC (2011) National Research Council committee for the update of the guide for the care and use of laboratory animals. Guide for the care and use of laboratory animals. 8th edition. Washington (DC): National Academies Press (US). https://www.ncbi.nlm.nih.gov/books/NBK54045
NTP (2018) National Toxicology Program approach to genomic dose-response modeling. NTP RR 5. Research Triangle Park. NC National Toxicology Program 5: 1–44. https://doi.org/10.22427/NTP-RR-5
Nyffeler J, Willis C, Lougee R, Richard A, Paul-Friedman K, Harrill JA (2020) Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol App Pharamacol 389:114876. https://doi.org/10.1016/j.taap.2019.114876
OECD (2013a) Test No. 210: Fish, early-life stage toxicity test. OECD guidelines for the testing of chemicals, section 2, OECD Publishing, Paris. https://doi.org/10.1787/9789264203785-en
OECD (2013b). Test No. 236: Fish embryo acute toxicity (FET) test. OECD guidelines for the testing of chemicals, section 2, OECD Publishing, Paris. https://doi.org/10.1787/9789264203709-en
Olker JH, Elonen CM, Pilli A, Anderson A, Kinziger B, Erickson S, Skopinski M, Pomplun A, LaLone CA, Russom CL, Hoff D (2022) The ECOTOXicology Knowledgebase: a curated database of ecologically relevant toxicity tests to support environmental research and risk assessment. Environ Toxicol Chem 41(6):1520–1539. https://doi.org/10.1002/etc.5324
Page-Lariviere F, Crump D, O’Brien JM (2019) Transcriptomic point-of-departure from short-term exposure studies are protective of chronic effects for fish exposed to estrogenic chemicals. Toxicol App Pharamacol 378:114634. https://doi.org/10.1016/j.taap.2019.114634
Paul-Friedman K, Gagne M, Loo LH, Karamertzanis P, Netzeva T, Sobanzki T, Franzosa JA, Richard AM, Lougee RR, Gissi A, Lee JYJ, Angrish M, Dorne JL, Foster S, Raffaele K, Bahadori T, Gwinn MR, Lambert J, Whelan M, Rasenberg M, Barton-Maclaren T, Thomas RS (2020) Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol Sci 173(1):202–225. https://doi.org/10.1093/toxsci/kfz201
Phillips JR, Svoboda DL, Tandon A, Patel S, Sedykh A, Mav D, Kuo B, Yauk CL, Yang L, Thomas RS, Gift JS, Davis JA, Olszyk L, Merrick BA, Paules RS, Parham F, Saddles T, Shah RR, Auerbach SS (2019) BMDExpress 2: enhanced transcriptomic dose-response analysis workflow. Bioinformatics 35(10):1780–1782. https://doi.org/10.1093/bioinformatics/bty878
R Core Team (2022) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Reardon AJF, Rowan-Carroll A, Ferguson SS, Leingartner K, Gagne R, Kuo B, Williams A, Lorusso L, Bourdon-Lacombe JA, Carrier R, Moffat I, Yauk CL, Atlas E (2021) Potency ranking of per- and polyfluoroalkyl substances using high-throughput transcriptomic analysis of human liver spheroids. Toxicol Sci 184(1):154–169. https://doi.org/10.1093/toxsci/kfab102
RStudio Team (2020) RStudio: Integrated development for R. RStudio, PBC, Boston, MA. www.rstudio.com.
Thomas RS, Allen B, Nong A, Yang L, Bermudez E, Clewell HJ, Andersen ME (2007) A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci 98(1):240–248. https://doi.org/10.1093/toxsci/kfm092
Thomas RS, Clewell HJ, Allen BC, Yang L, Healy E, Andersen ME (2012) Integrating pathway-based transcriptomic data into quantitative chemical risk assessment: a five chemical case study. Mutat Res 746:135–143. https://doi.org/10.1016/j.mrgentox.2012.01.007
Thomas RS, Wesselkamper SC, Wang NCY, Zhao QJ, Petersen DD, Lambert JC, Cote I, Yang L, Healy E, Black MB, Clewell HJ, Allen BC, Andersen ME (2013) Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci 134(1):180–194. https://doi.org/10.1093/toxsci/kft094
Thomas RS, Bahadori T, Buckley TJ, Cowden J, Deisenroth C, Dionisio KL, Frithsen JB, Grulke CM, Gwinn MR, Harrill JA, Higuchi M, Houck KA, Hughes MF, Hunter ES III, Isaacs KK, Judson RS, Knudsen TB, Lambert JC, Linnenbrink M, Martin TM, Newton SR, Padilla S, Patlewicz G, Paul-Friedman K, Phillips KA, Richard AM, Sams R, Shafer TJ, Setzer RW, Shah I, Simmons JE, Simmons SO, Singh A, Sobus JR, Strynar M, Swank A, Tornero-VAlex R, Ulrich EM, Villeneuve DL, Wambaugh JF, Wetmore BA, Williams AJ (2019) The next generation blueprint of computational toxicology at the US Environmental Protection Agency. Toxicol Sci 169(1):317–332. https://doi.org/10.1093/toxsci/kfz058
Tice R, Austin CP, Kavlock RJ, Bucher JR (2013) Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121(7). https://doi.org/10.1289/ehp.1205784
Uno T, Ishizuka M, Itakura T (2012) Cytochrome P450 (CYP) in fish. Environ Toxicol Pharmacol 34(1):1–13. https://doi.org/10.1016/j.etap.2012.02.004
USEPA (2016a) OCSPP 850.1075: Freshwater and saltwater fish acute toxicity test. www.regulations.gov/document/EPA-HQ-OPPT-2009-0154-0035.
USEPA (2016b) OCSPP 850.1400: Fish early life stage toxicity test. www.regulations.gov/document/EPA-HQ-OPPT-2009-0154-0033.
Vestel J, Caldwell DJ, Constantine L, D’Aco VJ, Davidson T, Dolan DG, Millard SP, Murray-Smith R, Parke NJ, Ryan JJ, Straub JO, Wilson P (2016) Use of acute and chronic ecotoxicity data in environmental risk assessment of pharmaceuticals. Environ Toxicol Chem 35(5):1201–1212. https://doi.org/10.1002/etc.3260
Villeneuve D, Le M, Hazemi M, Biales A, Bencic DC, Bush K, Flick R, Martinson J, Morshead M, Santana Rodriguez K, Vitense K, Flynn K (2023) Pilot testing and optimization of a larval fathead minnow high throughput transcriptomics assay. Cur Res Toxicol 4:100099. https://doi.org/10.1016/j.crtox.2022.100099
Wang M, Zhao Y, Zhang B (2015) Efficient test and visualization of multi-set intersections. Scient Reports 5:16923. https://doi.org/10.1038/srep16923
Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS, Richard AM (2017) The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminformatics 9:61. https://doi.org/10.1186/s13321-017-0247-6
Wiseman SB, He Y, Gamal-El Din M, Martin JW, Jones PD, Hecker M, Giesy JP (2013) Transcriptional responses of male fathead minnows exposed to oil sands process-affected water. Compar Biochem Physiol C Toxicol Pharmacol 157(2):227–235. https://doi.org/10.1016/j.cbpc.2012.12.002
Yang L, Allen BC, Thomas RS (2007) BMDExpress: a software tool for the benchmark dose analyses of genomic data. BMC Genomics 8:387. https://doi.org/10.1186/1471-2164-8-387
Yu SJ (2008) Detoxification mechanisms in insects. In: Capinera JL (ed) Encyclopedia of entomology. Springer, Dordrecht.
Acknowledgements
The authors thank M. Tapper and J. Harrill for comments on an earlier version of this manuscript, K. Jensen, J. Cavallin, L. Everett, L. Wehmas, and K. Vitense for technical discussions, and P. Schumann for assistance with uploading data to the Sequence Read Archive. This work is part of an Accelerating the Pace of Chemical Risk Assessment (APCRA) case study: “Transcriptomics-based points of departure for ecotoxicology—investigating the applicability of high-throughput transcriptomics data to inform quantitative hazard assessments for ecological species.” We thank our APCRA partners including C. Inglis, J. Prindiville, A. Nong, J. O’Brien, and F. Page-Lariviere.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Kevin Flynn contributed to Conceptualization, Methodology, Formal analysis, Investigation, Writing—Original Draft, Visualization, and Supervision. Michelle Le contributed to Methodology, Validation, Investigation, and Writing—Review & Editing. Monique Hazemi contributed to Methodology, Software, Formal analysis, Data Curation, and Writing—Review & Editing. Adam Biales contributed to Conceptualization, Resources, Writing—Review & Editing, and Supervision. David C. Bencic contributed to Methodology, Investigation, Resources, and Writing—Review & Editing. Brett R. Blackwell contributed to Methodology, Validation, Writing—Review & Editing, and Supervision. Kendra Bush contributed to Methodology, Validation, Investigation, and Writing—Review & Editing. Robert Flick contributed to Methodology, Investigation, Resources, and Writing—Review & Editing. John X. Hoang contributed to Investigation. John Martinson contributed to Software and Data Curation. Mackenzie Morshead contributed to Investigation. Kelvin Santana Rodriguez contributed to Investigation. Emma Stacy contributed to Methodology, Validation, Investigation, and Writing—Review & Editing. Daniel L. Villeneuve contributed to Conceptualization, Methodology, Formal analysis, Investigation, Writing—Original Draft, Visualization, and Supervision.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Disclaimer
The contents of this manuscript neither constitute nor necessarily reflect US EPA policy. Mention of trade names or commercial products does not constitute endorsement or recommendation of use.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Flynn, K., Le, M., Hazemi, M. et al. Comparing Transcriptomic Points of Departure to Apical Effect Concentrations For Larval Fathead Minnow Exposed to Chemicals with Four Different Modes Of Action. Arch Environ Contam Toxicol (2024). https://doi.org/10.1007/s00244-024-01064-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00244-024-01064-y