Abstract
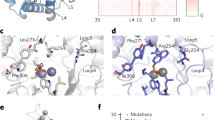
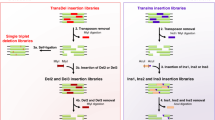
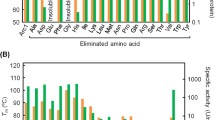
The study of evolution is important to understand biological phenomena. During evolutionary processes, genetic changes confer amino acid substitutions in proteins, resulting in new or improved functions. Unfortunately, most mutations destabilize proteins. Thus, protein stability is a significant factor in evolution; however, its role remains unclear. Here, we simply and directly explored the association between protein activity and stability in random mutant libraries to elucidate the role of protein stability in evolutionary processes. In the first random mutation of an esterase from Sulfolobus tokodaii, approximately 20% of the variants displayed higher activity than wild-type protein (i.e., 20% evolvability). During evolutionary processes, the evolvability depended on the stability of template proteins, indicating that protein evolution is potentially governed by protein stability. Furthermore, decreased activity could be recovered during evolution by maintaining the stability of variants. The results suggest that protein sequence space for its evolution is able to expand during nearly neutral evolution where mutations are slightly deleterious for activity but rarely fatal for stability. Molecular evolution is a crucial phenomenon that has continued since the birth of life on earth, and mechanism underlying it is simple; therefore, this could be demonstrated by our simple experiments. These findings also can be applied to protein engineering.





Similar content being viewed by others
References
Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS (2005) The ‘evolvability’ of promiscuous protein functions. Nat Genet 37:73–76
Akashi H, Osada N, Ohta T (2012) Weak selection and protein evolution. Genet 192:15–31
Angkawidjaja C, Koga Y, Takano K, Kanaya S (2012) Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobus tokodaii. FEBS J 279:3071–3084
Ashenberg O, Gong LI, Bloom JD (2013) Mutational effects on stability are largely conserved during protein evolution. Proc Natl Acad Sci USA 110:21071–21076
Bastolla U, Dehouck Y, Echave J (2017) What evolution tells us about protein physics, and protein physics tells us about evolution. Curr Opin Struct Biol 42:59–66
Berezovsky IN, Shakhnovich EI (2005) Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci USA 102:12742–12747
Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS (2006) Robustness–epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444(7121):929
Bershtein S, Goldin K, Tawfik DS (2008) Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol 379:1029–1044
Bershtein S, Serohijos AW, Shakhnovich EI (2017) Bridging the physical scales in evolutionary biology: from protein sequence space to fitness of organisms and populations. Curr Opin Struct Biol 42:31–40
Blaber M, Zhang XJ, Matthews BW (1993) Structural basis of amino acid alpha helix propensity. Science 260:1637–1640
Bloom JD, Labthavikul ST, Otey CR, Arnold FH (2006) Protein stability promotes evolvability. Proc Natl Acad Sci USA 103:5869–5874
Bloom JD, Raval A, Wilke CO (2007) Thermodynamics of neutral protein evolution. Genet 175:255–266
Bogumil D, Dagan T (2012) Cumulative impact of chaperone-mediated folding on genome evolution. Biochemistry 51:9941–9953
Caetano-Anollés G, Mittenthal J (2010) Exploring the interplay of stability and function in protein evolution. Bioessays 32:655–658
Carneiro M, Hartl DL (2010) Adaptive landscapes and protein evolution. Proc Natl Acad Sci USA 107:1747–1751
Darwin CR (1859) On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray, London
DePristo MA, Weinreich DM, Hartl DL (2005) Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6:678–687
Draghi JA, Parsons TL, Wagner GP, Plotkin JB (2010) Mutational robustness can facilitate adaptation. Nature 463:353–355
Elena SF, Sanjuán R (2008) The effect of genetic robustness on evolvability in digital organisms. BMC Evol Biol 8:284
Funahashi J, Takano K, Yamagata Y, Yutani K (1999) Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme. Protein Eng 12:841–850
Godoy-Ruiz R, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM (2004) Relation between protein stability, evolution and structure, as probed by carboxylic acid mutations. J Mol Biol 336:313–318
Gong LI, Suchard MA, Bloom JD (2013) Stability-mediated epistasis constrains the evolution of an influenza protein. eLife 2:e00631
Kaltenbach M, Tokuriki N (2014) Dynamics and constraints of enzyme evolution. J Exp Zool B 322:468–487
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626
Mack KL, Shorter J (2016) Engineering and evolution of molecular chaperones and protein disaggregases with enhanced activity. Front Mol Biosci 3:8
Magliery TJ, Lavinder JJ, Sullivan BJ (2011) Protein stability by number: high-throughput and statistical approaches to one of protein science’s most difficult problems. Curr Opin Chem Biol 15:443–451
Matthews BW, Nicholson H, Becktel WJ (1987) Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci USA 84:6663–6667
Miton CM, Tokuriki N (2016) How mutational epistasis impairs predictability in protein evolution and design. Protein Sci 25:1260–1272
Mizuguchi K, Sele M, Cubellis MV (2007) Environment specific substitution tables for thermophilic proteins. BMC Bioinform 8:S15
Mukaiyama A, Haruki M, Ota M, Koga Y, Takano K, Kanaya S (2006) A hyperthermophilic protein acquires function at the cost of stability. Biochemistry 45:12673–12679
Nelson KE et al (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329
Ohta T (1973) Slightly deleterious mutant substitutions in evolution. Nature 246:96–98
Ohta T (1992) The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23:263–286
Okada J, Okamoto T, Mukaiyama A, Tadokoro T, You DJ, Chon H, Koga Y, Takano K, Kanaya S (2010) Evolution and thermodynamics of the slow unfolding of hyperstable monomeric proteins. BMC Evol Biol 10:207
Philip AF, Kumauchi M, Hoff WD (2010) Robustness and evolvability in the functional anatomy of a PER-ARNT-SIM (PAS) domain. Proc Natl Acad Sci USA 107:17986–17991
Romero PA, Arnold FH (2009) Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol 10:866–876
Serrano L, Neira JL, Sancho J, Fersht AR (1992) Effect of alanine versus glycine in alpha-helices on protein stability. Nature 356:453–455
Sikosek T, Chen HS (2014) Biophysics of protein evolution and evolutionary protein biophysics. J R Soc Interface 11:20140419
Socha RD, Tokuriki N (2013) Modulating protein stability—directed evolution strategies for improved protein function. FEBS J 280:5582–5595
Son SY, Ma J, Kondou Y, Yoshimura M, Yamashita E, Tsukihara T (2008) Structure of human monoamine oxidase A at 2.2-Å resolution: the control of opening the entry for substrates/inhibitors. Proc Natl Acad Sci USA 105:5739–5744
Starr TN, Thornton JW (2016) Epistasis in protein evolution. Protein Sci 25:1204–1218
Steinberg B, Ostermeier M (2016a) Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway. J Mol Biol 428:2730–2743
Steinberg B, Ostermeier M (2016b) Environmental changes bridge evolutionary valleys. Sci Adv 2:e1500921
Suzuki Y, Miyamoto K, Ohta H (2004) A novel thermostable esterase from the thermoacidophilic archaeon Sulfolobus tokodaii strain 7. FEMS Microbiol Lett 236:97–102
Takano K, Ogasahara K, Kaneda H, Yamagata Y, Fujii S, Kanaya E, Kikuchi M, Oobatake M, Yutani K (1995) Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants. J Mol Biol 254:62–76
Takano K, Yamagata Y, Kubota M, Funahashi J, Fujii S, Yutani K (1999) Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser → Ala mutants. Biochemistry 38:6623–6629
Takano K, Tsuchimori K, Yamagata Y, Yutani K (2000) Contribution of salt bridges near the surface of a protein to the conformational stability. Biochemistry 39:12375–12381
Takano K, Scholtz JM, Sacchettini JC, Pace CN (2003) The contribution of polar group burial to protein stability is strongly context-dependent. J Biol Chem 278:31790–31795
Takano K, Aoi A, Koga Y, Kanaya S (2013) Evolvability of thermophilic proteins from Archaea and Bacteria. Biochemistry 52:4774–4780
Tokuriki N, Tawfik DS (2009a) Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19:596–604
Tokuriki N, Tawfik DS (2009b) Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459:668–673
Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS (2007) The stability effects of protein mutations appear to be universally distributed. J Mol Biol 369:1318–1332
Tokuriki N, Stricher F, Serrano L, Tawfik DS (2008) How protein stability and new functions trade off. PLoS Comput Biol 4:e1000002
Wang X, Minasov G, Shoichet BK (2002) Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol 320:85–95
Worth CL, Gong S, Blundell TL (2009) Structural and functional constraints in the evolution of protein families. Nat Rev Mol Cell Biol 10:709–720
Wyganowski KT, Kaltenbach M, Tokuriki N (2013) GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol 425:3403–3414
Yang G, Hong N, Baier F, Jackson CJ, Tokuriki N (2016) Conformational tinkering drives evolution of a promiscuous activity through indirect mutational effects. Biochemistry 55:4583–4593
Zeldovich KB, Chen P, Shakhnovich EI (2007a) Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA 104:16152–16157
Zeldovich KB, Chen P, Shakhnovich BE, Shakhnovich EI (2007b) A first-principles model of early evolution: emergence of gene families, species, and preferred protein folds. PLoS Comput Biol 3:e139
Acknowledgements
This work was supported in part by grants from the Japan Society for the Promotion of Science to KT (KAKENHI Nos. 25440194 and 17K07368), by a Research Encouragement Project from the Academic Promotion Fund of Kyoto Prefectural University to RK (2016), and by a Young Researcher Development Support Project from the Kyoto Prefectural Public University Corporation to RK (2017).
Author information
Authors and Affiliations
Contributions
KT conceived and supervised the experiments. RK performed the experiments. RK and KT wrote the paper. SS helped in interpretation of data and discussion of results. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Rights and permissions
About this article
Cite this article
Kurahashi, R., Sano, S. & Takano, K. Protein Evolution is Potentially Governed by Protein Stability: Directed Evolution of an Esterase from the Hyperthermophilic Archaeon Sulfolobus tokodaii. J Mol Evol 86, 283–292 (2018). https://doi.org/10.1007/s00239-018-9843-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-018-9843-y