Abstract
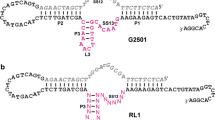
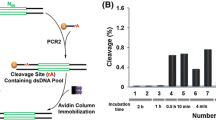
The leadzyme refers to a small ribozyme that cleaves a RNA substrate in the presence of Pb2+. In an optimized form, the enzyme strand contains only two unpaired nucleotides. Most RNA-cleaving DNAzymes are much longer. Two classical Pb2+-dependent DNAzymes, 8–17 and GR5, both contain around 15 nucleotides in the enzyme loop. This is also the size of most RNA-cleaving DNAzymes that use other metal ions for their activity. Such large enzyme loops make spectroscopic characterization difficult and so far no high-resolution structural information is available for active DNAzymes. The goal of this work is to search for DNAzymes with smaller enzyme loops. A simple replacement of the ribonucleotides in the leadzyme by deoxyribonucleotides failed to produce an active enzyme. A Pb2+-dependent in vitro selection combined with deep sequencing was then performed. After sequence alignment and DNA folding, a new DNAzyme named PbE22 was identified, which contains only 5 nucleotides in the enzyme catalytic loop. The biochemical characteristics of PbE22 were compared with those of the leadzyme and the two classical Pb2+-dependent DNAzymes. The rate of PbE22 rises with increase in Pb2+ concentration, being 1.7 h−1 in the presence of 100 μM Pb2+ and reaching 3.5 h−1 at 500 µM Pb2+. The log of PbE22 rate rises linearly in a pH-dependent fashion (20 µM Pb2+) with a slope of 0.74. In addition, many other abundant sequences in the final library were studied. These sequences are quite varied in length and nucleotide composition, but some contain a few conserved nucleotides consistent with the GR5 structure. Interestingly, some sequences are active with Pb2+ but none of them were active with even 50 mM Mg2+, which is reminiscent of the difference between the GR5 and 8–17 DNAzymes.






Similar content being viewed by others
References
Ameta S, Winz M-L, Previti C, Jäschke A (2014) Next-generation sequencing reveals how RNA catalysts evolve from random space. Nucleic Acids Res 42:1303–1310
Breaker RR, Joyce GF (1994) A DNA enzyme that cleaves RNA. Chem Biol 1:223–229
Brown RS, Dewan JC, Klug A (1985) Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry 24:4785–4801
Brown AK, Li J, Pavot CMB, Lu Y (2003) A lead-dependent DNAzyme with a two-step mechanism. Biochemistry 42:7152–7161
Brown AK, Liu JW, He Y, Lu Y (2009) Biochemical characterization of a uranyl ion-specific DNAzyme. ChemBioChem 10:486–492
Chartrand P, Usman N, Cedergren R (1997) Effect of structural modifications on the activity of the leadzyme. Biochemistry 36:3145–3150
Cruz RPG, Withers JB, Li Y (2004) Dinucleotide junction cleavage versatility of 8–17 deoxyribozyme. Chem Biol 11:57–67
Dimroth K, Witzel H, Hulsen W, Mirbach H (1959) Uber die hydrolyse von ribonucleinsauren in gegenwart von metallhydroxyden. Annalen Der Chemie-Justus Liebig 620:94–108
Farkas WR (1968) Depolymerization of ribonucleic acid by plumbous ion. Biochimica et biophysica acta (BBA)—nucleic acids and protein. Synthesis 155:401–409
Hoogstraten CG, Legault P, Pardi A (1998) NMR solution structure of the lead-dependent ribozyme: evidence for dynamics in RNA catalysis. J Mol Biol 284:337–350
Hoogstraten CG, Wank JR, Pardi A (2000) Active site dynamics in the lead-dependent ribozyme. Biochemistry 39:9951–9958
Huang P-JJ, Liu J (2014) Two Pb2+-specific dnazymes with opposite trends in split-site-dependent activity. Chem Commun 50:4442–4444
Huang P-JJ, Lin J, Cao J, Vazin M, Liu J (2014a) Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal Chem 86:1816–1821
Huang P-JJ, Vazin M, Liu J (2014b) In vitro selection of a new lanthanide-dependent DNAzyme for ratiometric sensing lanthanides. Anal Chem 86:9993–9999
Kadakkuzha BM, Zhao L, Xia T (2009) Conformational distribution and ultrafast base dynamics of leadzyme. Biochemistry 48:3807–3809
Kim HK, Rasnik I, Liu JW, Ha TJ, Lu Y (2007) Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat Chem Biol 3:762–768
Lan T, Furuya K, Lu Y (2010) A highly selective lead sensor based on a classic lead DNAzyme. Chem Commun 46:3896–3898
Legault P, Hoogstraten CG, Metlitzky E, Pardi A (1998) Order, dynamics and metal-binding in the lead-dependent ribozyme. J Mol Biol 284:325–335
Li Y, Breaker RR (1999) Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc 121:5364–5372
Li J, Lu Y (2000) A highly sensitive and selective catalytic DNA biosensor for lead ions. J Am Chem Soc 122:10466–10467
Li J, Zheng W, Kwon AH, Lu Y (2000) In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res 28:481–488
Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y (2007) A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci USA 104:2056–2061
Liu J, Cao Z, Lu Y (2009) Functional nucleic acid sensors. Chem Rev 109:1948–1998
Lu Y (2002) New transition metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem Eur J 8:4588–4596
Majerfeld I, Chocholousova J, Malaiya V, Widmann J, McDonald D, Reeder J, Iyer M, Illangasekare M, Yarus M, Knight R (2010) Nucleotides that are essential but not conserved; a sufficient l-tryptophan site in RNA. RNA 16:1915–1924
Mao Y, Liu M, Tram K, Gu J, Salena BJ, Jiang Y, Li Y (2015) Optimal DNA templates for rolling circle amplification revealed by in vitro selection. Chem Eur J 21:8069–8074
Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF (1999) Crystal structure of an 82-nucleotide RNA–DNA complex formed by the 10-23 DNA enzyme. Nat Struct Biol 6:151–156
Ohmichi T, Okumoto Y, Sugimoto N (1998) Effect of substrate RNA sequence on the cleavage reaction by a short ribozyme. Nucleic Acids Res 26:5655–5661
Pan T, Uhlenbeck OC (1992a) In vitro selection of rnas that undergo autolytic cleavage with Pb(II). Biochemistry 31:3887–3895
Pan T, Uhlenbeck OC (1992b) A small metalloribozyme with a two-step mechanism. Nature 358:560–563
Pan T, Dichtl B, Uhlenbeck OC (1994) Properties of an in vitro selected Pb2+ cleavage motif. Biochemistry 33:9561–9565
Peracchi A, Bonaccio M, Clerici M (2005) A mutational analysis of the 8-17 deoxyribozyme core. J Mol Biol 352:783–794
Pitt JN, Ferré-D’Amaré AR (2010) Rapid construction of empirical RNA fitness landscapes. Science 330:376–379
Santoro SW, Joyce GF (1997) A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA 94:4262–4266
Schlosser K, Li YF (2009) Biologically inspired synthetic enzymes made from DNA. Chem Biol 16:311–322
Schlosser K, Li Y (2010) A versatile endoribonuclease mimic made of DNA: characteristics and applications of the 8-17 RNA-cleaving DNAzyme. ChemBioChem 11:866–879
Schlosser K, Gu J, Lam JCF, Li YF (2008a) In vitro selection of small RNA-cleaving deoxyribozymes that cleave pyrimidine-pyrimidine junctions. Nucleic Acids Res 36:4768–4777
Schlosser K, Gu J, Sule L, Li YF (2008b) Sequence-function relationships provide new insight into the cleavage site selectivity of the 8-17 RNA-cleaving deoxyribozyme. Nucleic Acids Res 36:1472–1481
Sigel RKO, Pyle AM (2007) Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem Rev 107:97
Wang B, Cao L, Chiuman W, Li Y, Xi Z (2010) Probing the function of nucleotides in the catalytic cores of the 8-17 and 10-23 dnazymes by abasic nucleotide and C3 spacer substitutions. Biochemistry 49:7553–7562
Ward WL, Plakos K, DeRose VJ (2014) Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem Rev 114:4318–4342
Wedekind JE, McKay DB (1999) Crystal structure of a lead-dependent ribozyme revealing metal binding sites revelant to catalysis. Nat Struct Biol 6:261–268
Wedekind JE, McKay DB (2003) Crystal structure of the leadzyme at 1.8 Å resolution: metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry 42:9554–9563
Werner C, Krebs B, Keith G, Dirheimer G (1976) Specific cleavages of pure tRNAs by plumbous ions. Biochim Biophys Acta 432:161–175
Winterme W, Zachau HG (1973) Mg2+-catalyzed specific splitting of transfer-RNA. Biochim Biophys Acta 299:82–90
Zhang X-B, Kong R-M, Lu Y (2011) Metal ion sensors based on DNAzymes and related DNA molecules. Annu Rev Anal Chem 4:105–128
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
Funding for this work was from the University of Waterloo, Ontario Ministry of Research & Innovation, and the Natural Sciences and Engineering Research Council (NSERC) of Canada (Discovery Grant and Strategic Project Grant).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saran, R., Chen, Q. & Liu, J. Searching for a DNAzyme Version of the Leadzyme. J Mol Evol 81, 235–244 (2015). https://doi.org/10.1007/s00239-015-9702-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-015-9702-z