Abstract
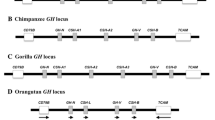
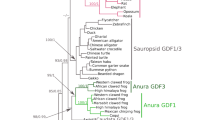
The relaxin (RLN) and insulin-like (INSL) gene family is a group of genes involved in a variety of physiological roles that includes bone formation, testicular descent, trophoblast development, and cell differentiation. This family appears to have expanded in vertebrates relative to non-vertebrate chordates, but the relative contribution of whole genome duplications (WGDs) and tandem duplications to the observed diversity of genes is still an open question. Results from our comparative analyses favor a model of divergence post vertebrate WGDs in which a single-copy progenitor found in the last common ancestor of vertebrates experienced two rounds of WGDs before the functional differentiation that gave rise to the RLN and INSL genes. One of the resulting paralogs was subsequently lost, resulting in three proto-RLN/INSL genes on three separate chromosomes. Subsequent rounds of tandem gene duplication and divergence originated the set of paralogs found on a given cluster in extant vertebrates. Our study supports the hypothesis that differentiation of the RLN and INSL genes took place independently in each RLN/INSL cluster after the two WGDs during the evolutionary history of vertebrates. In addition, we show that INSL4 represents a relatively old gene that has been apparently lost independently in all Euarchontoglires other than apes and Old World monkeys, and that RLN2 derives from an ape-specific duplication.




Similar content being viewed by others
References
Bathgate RAD, Samuel CS, Burazin TCD, Gundlach AL, Tregear GW (2003) Relaxin: new peptides, receptors and novel actions. Trends Endocrinol Metab 14:207–213
Bieche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo J-L, Olivi M, Fausser J-L, Evain-Brion D, Vidaud M (2003) Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod 68:1422–1429
Chan SJ, Steiner DF (2000) Insulin through the ages: phylogeny of a growth promoting and metabolic regulatory hormone. Am Zool 40:222–231
de Pablo F, de la Rosa EJ (1995) The developing CNS: a scenario for the action of proinsulin, insulin and insulin-like growth factors. Trends Neurosci 18:143–150
Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3:314
Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S (2005) ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res 15:330–340
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Good-Avila SV, Yegorov S, Harron S, Bogerd J, Glen P, Ozon J, Wilson BC (2009) Relaxin gene family in teleosts: phylogeny, syntenic mapping, selective constraint, and expression analysis. BMC Evol Biol 9:293
Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP (1998) Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol 9:585–598
Hahn MW (2009) Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100:605–617
Hsu SYT (2003) New insights into the evolution of the relaxin–LGR signaling system. Trends Endocrinol Metab 14:303–309
Jobb G, Haeseler AV, Strimmer K (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4:18
Katoh K, Asimenos G, Toh H (2009) Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64
Kuraku S, Meyer A, Kuratani S (2009) Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after. Mol Biol Evol 26:47–59
Lassmann T, Sonnhammer ELL (2005) Automatic assessment of alignment quality. Nucleic Acids Res 33:7120–7128
Lassmann T, Frings O, Sonnhammer ELL (2009) Kalign2: high performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res 37:858–865
Lynch M (2007) The origins of genome architecture. Sinauer Associates, Sunderland, MA
Meyer A, Schartl M (1999) Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 11:699–704
Nagamatsu S, Chan SJ, Falkmer S, Steiner DF (1991) Evolution of the insulin gene superfamily. Sequence of a preproinsulin-like growth factor cDNA from the Atlantic hagfish. J Biol Chem 266:2397–2402
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217
Ohno S (1970) Evolution by gene duplication. Springer-Verlag, New York
Olinski RP, Dahlberg C, Thorndyke M, Hallbook F (2006a) Three insulin–relaxin-like genes in Ciona intestinalis. Peptides 27:2535–2546
Olinski RP, Lundin L-G, Hallbook F (2006b) Conserved synteny between the Ciona genome and human paralogons identifies large duplication events in the molecular evolution of the insulin–relaxin gene family. Mol Biol Evol 23:10–22
Park J-I, Semyonov J, Chang CL, Yi W, Warren W, Hsu SYT (2008a) Origin of INSL3-mediated testicular descent in therian mammals. Genome Res 18:974–985
Park J-I, Semyonov J, Yi W, Chang CL, Hsu SYT (2008b) Regulation of receptor signaling by relaxin A chain motifs: derivation of pan-specific and LGR7-specific human relaxin analogs. J Biol Chem 283:32099–32109
Reinecke M, Collet C (1998) The phylogeny of the insulin-like growth factors. Int Rev Cytol 183:1–94
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sherwood OD (2004) Relaxin’s physiological roles and other diverse actions. Endocr Rev 25:205–234
Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508
Steiper ME, Young NM (2009) Primates (Primates). In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, Oxford, pp 482–486
Subramanian AR, Kaufmann M, Morgenstern B (2008) DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol Biol 3:6
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:609–612
Tatusova TA, Madden TL (1999) BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174:247–250
Wentworth BM, Schaefer IM, Villa-Komaroff L, Chirgwin JM (1986) Characterization of the two nonallelic genes encoding mouse preproinsulin. J Mol Evol 23:305–312
Wilkinson TN, Speed TP, Tregear GW, Bathgate RAD (2005) Evolution of the relaxin-like peptide family. BMC Evol Biol 5:14
Zhang J (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18:292–298
Acknowledgments
This work was funded by grants to JCO from the Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 11080181), Programa Bicentenario de Ciencia y Tecnología (PSD89), and the Oliver Pearson Award from the American Society of Mammalogists (ASM). The authors also thank Dominique Alò, Amy Runck and Zachary A. Cheviron for critical comments, and Yves Van de Peer and two anonymous reviewers for helpful suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hoffmann, F.G., Opazo, J.C. Evolution of the Relaxin/Insulin-like Gene Family in Placental Mammals: Implications for Its Early Evolution. J Mol Evol 72, 72–79 (2011). https://doi.org/10.1007/s00239-010-9403-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9403-6