Abstract
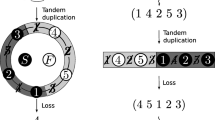
Asymmetric compositional and mutation bias between the two strands occurs in mitochondrial genomes, and an asymmetric mechanism of mtDNA replication is a potential source of this bias. Some evidence indicates that during replication the heavy strand is subject to a gradient of time spent in a single-stranded state (D ssH) and a gradient of mutational damage. The nucleotide composition bias among genes varies with D ssH. Consequently, partial genome duplications (PGD) will alter the skew for genes located downstream of the duplication, relatively to nascent light strand synthesis, and in the same way, gene rearrangements (GRr) will affect genes by changing their skews. We examined cases where there had been PGD or GRr and determined whether this left a trace in the form of unusual patterns of base composition. We compared the skew of genes differently located on the mtDNA genome of previously published whole mtDNA genomes from amphibians, a group that shows considerable levels of both GRr and PGD. After observing a significant correlation between AT and GC skew with D ssH at fourfold redundant sites, we ran our analysis and detected 31.3% of the species with GRr and/or PGD. By comparing the nucleotide composition at fourfold redundant sites in normal and “abnormal” species, we found that A/C variation occurs and is associated with GRr/PGD. These results show that by analyzing the nucleotide skews of only three genes, it may be possible to predict some mitochondrial GRr and/or PGD without knowing the complete mtDNA genome sequence.



Similar content being viewed by others
References
Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Asakawa S, Kumazawa Y, Araki T, Himeno H, Miura K, Watanabe K (1991) Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol 32:511–520
Ballard JW, Rand DM (2005) The population biology of mitochondrial DNA and its phylogenetics implications. Annu Rev Ecol Syst 36:621–642
Bielawski JP, Gold JR (2002) Mutation patterns of mitochondrial H- and L-strand DNA in closely related Cyprinid fishes. Genetics 161:1589–1597
Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA (2005) Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev 19:2466–2476
Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28:693–705
Faith JJ, Pollock DD (2003) Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics 165:735–745
Frank AC, Lobry JR (1999) Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238:65–77
Holt IJ, Lorimer HE, Jacobs HT (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100:515–524
Jermiin LS, Graur D, Crozier H (1995) Evidence from analyses of intergenic regions for strand-specific directional mutation pressure in metazoan mitochondrial DNA. Mol Biol Evol 12:558–563
Krishnan NM, Raina SZ, Pollock DD (2004a) Analysis of among-site variation in substitution patterns. Biol Proced Online 6:180–188
Krishnan NM, Seligmann H, Raina SZ, Pollock DD (2004b) Detecting gradients of asymmetry in site-specific substitutions in mitochondrial genomes. DNA Cell Biol 23:707–714
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715
Lobry JR (1995) Properties of a general model of DNA evolution under no-strand-bias conditions. J Mol Evol 40:326–330 (Erratum: J Mol Evol 41:680)
Lobry JR (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13:660–665
Macey JR, Shulte II JA, Larson A, Papenfuss TJ (1998) Tandem duplication via light-strand synthesis may provide a precursor for mitochondrial genomic rearrangement. Mol Biol Evol 15:71–75
Mizi A, Zouros E, Moschonas N, Rodakis GC (2005) The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Mol Biol Evol 22:952–967
Mueller RL, Boore JL (2005) Molecular mechanisms of extensive mitochondrial gene rearrangement in Plethodontid salamanders. Mol Biol Evol 22:2104–2112
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Radloff R, Bauer W, Vinograd J (1967) A dye-buoyant density method for the detection and isolation of closed circular DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci USA 57:1514–1521
Raina SZ, Faith JJ, Disotell TR, Seligmann H, Stewart CB, Pollock DD (2005) Evolution of base-substitution gradients in primate mitochondrial genomes. Genome Res 15:665–673
Reyes A, Gissi C, Pesole G, Saccone C (1998) Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol 15:957–966
Sueoka N (1995) Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol 40:318–325
Tanaka M, Ozawa T (1994) Strand asymmetry in human mitochondrial DNA mutations. Genomics 22:327–335
Yang MY, Bowmaker M, Reyes A, Vergani L, Paolo A, Gringeri E, Jacobs HT, Holt IJ (2002) Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111:495–505
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia (FCT) grants POCI/61946/2004 (to D.J.H), SFRH/BD/11377/2002 (to E.F.), and POCI/BIA-BDE/61946/2004 (to M.M.F.). We thank the two reviewers for their constructive comments on early drafts of the manuscript and D. Pollock for his extensive editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. David Pollock]
Rights and permissions
About this article
Cite this article
Fonseca, M.M., Froufe, E. & James Harris, D. Mitochondrial Gene Rearrangements and Partial Genome Duplications Detected by Multigene Asymmetric Compositional Bias Analysis. J Mol Evol 63, 654–661 (2006). https://doi.org/10.1007/s00239-005-0242-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0242-9