Abstract
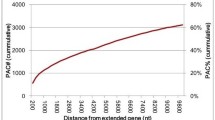
We identified 411 processed sequences in the Arabidopsis thaliana genome based on the fact that they have lost their intron(s) and have a length that is at least 95% of the length of the gene that gave rise to them. These sequences were generated by 230 different genes and clearly originated from retrotranspositons events because most of them (91%) have a poly(A)-tail. They are composed of 376 sequences with frame shifts and/or premature stop codons (processed pseudogenes) and 35 sequences without disablements (processed genes). Eleven of these processed genes are likely functional retrotransposed genes because they have low Ka/Ks ratios and high Ks values, and their sequences match numerous Arabidopsis ESTs. Processed sequences are mostly randomly distributed in the Arabidopsis genome and their rate of accumulation has steadily been decreasing since it peaked some 50 MYA. In contrast with the situation observed in mammals, the processed sequences found in the Arabidopsis genome originate from genes with high copy numbers and not from highly expressed genes. The patterns of spontaneous mutations in Arabidopsis are slightly different than those of mammals but are similar to those observed in Drosophila. This suggests that methylated cytosine deamination is less frequent in Arabidopsis than in mammals.




Similar content being viewed by others
References
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Brosius J (1999) RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238:115–134
Brosius J, Gould SJ (1992) On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.” Proc Natl Acad Sci USA 89:10706–10710
Cho S, Jin SW, Cohen A, Ellis RE (2004). A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res 14:1207–1220
Coulondre C, Miller JH, Farabaugh PJ, Gilbert W (1978) Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775–780
Drouin G, Dover GA (1987) A plant processed pseudogene. Nature 328:557–558
Drouin G, Moniz de Sá M (1997) Loss of introns in the pollen-specific actin gene subfamily members of potato and tomato. J Mol Evol 45:509–513
Fink GR (1987) Pseudogenes in yeast? Cell 49:5–6
Gojobori T, Li W-H, Graur D (1982) Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol 18:360–369
Gonçalves I, Duret L, Mouchiroud D (2000) Nature and structure of human genes that generate retropseudognes. Genome Res 10:672–678
Graur D, Li W-H (1999) Fundamentals of molecular evolution, 2nd ed. Sinauer Associates, Sunderland, MA
Gruenbaum Y, Naveh-Many T, Cedar H, Razin A (1981) Sequence specificity of methylation in higher plant DNA. Nature 292:860–862
Harrison PM, Echols N, Gerstein MB (2001) Digging for dead genes: an analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res 29:818–830
Harrison PM, Kumar A, Lan N, Echols N, Snyder M, Gerstein M (2002) A small reservoir of disabled ORF’s in the yeast genome and its implications for the dynamics of proteome evolution. J Mol Biol 316:409–419
Harrison PM, Milburn D, Zhang Z, Bertone P, Gerstein M (2003) Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res 31:1033–1037
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotides sequences. J Mol Evol 16:111–120
Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A (2003) An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423:91–96
Kvarnheden A, Tandre K, Engstrom P (1995) A cdc2 homologue and closely related processed retropseudogenes from Norway spruce. Plant Mol Biol 27:391–403
Li W-H (1993) Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99
Li W-H, Gojobori T, Nei M (1981) Pseudogenes as a paradigm of neutral evolution. Nature 292:237–239
Li W-H, Wu CI, Luo CC (1984) Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J Mol Evol 21:58–71
McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB (1996) Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142:587–602
Mladek C, Guger K, Hauser MT (2003) Identification and characterization of the ARIADNE gene family in Arabidopsis. A group of putative E3 ligases. Plant Physiol 131:27–40
Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, Smith CD, Tupy JL, Whitfied EJ, Bayraktaroglu L, Berman BP, Bettencourt BR, Celniker SE, de Grey AD, Drysdale RA, Harris NL, Richter J, Russo S, Schroeder AJ, Shu SQ, Stapleton M, Yamada C, Ashburner M, Gelbart WM, Rubin GM, Lewis SE (2002) Annotation of the Drosophila melanogaster euchromatic genome:a systematic review. Genome Biol 3:research0083
Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N (2003) Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol 4:R74
Ophir R, Graur D (1997) Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene 205:191–202
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Petrov DA, Hartl DL (1999) Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc Natl Acad Sci USA 96:1475–1479
Razin A, Riggs AD (1980) DNA methylation and gene function. Science 210:604–610
Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes HW, Mayer KF (2002) MIPS Arabidopsis thaliana Database (MAtDB):an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res 30:91–93
Torrents D, Suyama M, Zdobnov E, Bork P (2003) A genome-wide survey of human pseudogenes. Genome Res 13:2259–2567
Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A (1982) The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett 146:148–152
Vanin EF (1985) Processed pseudogenes: characteristics and evolution. Annu Rev Genet 19:253–272
Weiner AM, Deininger PL, Efstratiadis A (1986) Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem 55:631–661
Zar JH (1999) Biostatistical analysis, 4th ed. Prentice Hall, Upper Saddle River, NJ
Zhang Z, Gerstein M (2003) Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res 31:5338–5348
Zhang Z, Harrison P, Gerstein M (2002) Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res 12:1466–1482
Zhang Z, Harrison PM, Liu L, Gerstein M (2003) Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res 13:2541–2558
Zhang Z, Carriero N, Gerstein M (2004) Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet 20:62–67
Acknowledgments
We thank Robert Morris for his help with writing several PERL scripts. We also thank the Associate Editor and the two anonymous referees for their constructive comments on a previous version of the manuscript. This work was supported by a Discovery Grant from the National Science and Engineering Research Council of Canada to G.D.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Juergen Brosius]
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Benovoy, D., Drouin, G. Processed Pseudogenes, Processed Genes, and Spontaneous Mutations in the Arabidopsis Genome. J Mol Evol 62, 511–522 (2006). https://doi.org/10.1007/s00239-005-0045-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0045-z