Abstract
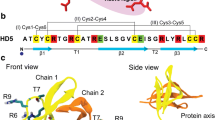
Plant defensins are a part of the innate immune system of plants that acts against a broad range of pathogens. Many plant defensins, including pine defensins, show strong antifungal activity that is associated with their ability to penetrate into the fungal cell membrane. However, the exact molecular mechanism of their action remains poorly defined. To obtain insight into the mechanism of protein–membrane interaction, we applied a coarse-grained molecular dynamics simulation to study the interaction of pine defensin with two model membranes: the first consisted of zwitterion-neutral POPC molecules and the second was composed of combined anionic POPG and POPC. The simulations show that defensin does not form stable complexes with the neutral membrane but does interact with the combined POPG/POPC membrane. In the latter case, defensin attaches to the membrane surface by interacting with lipid polar heads without deep penetration into the hydrophobic tail zone. Electrostatic interactions are a driving force of the complex formation, which determines the orientation of the protein relative to the bilayer surface. Two favorable orientations of defensin are detected where the defensin molecule orients either perpendicular or parallel to the membrane plane. Being positively charged, pine defensin induces changes in the lipid distribution along the membrane, resulting in the formation of zones with different electrostatic potentials that can cause deformation or distortion of the membrane. Pine defensin is a representative of plant defensins, and hence the results of this study can be applied to other members of the family.











Similar content being viewed by others
References
Aerts AM, François IE, Bammens L, Cammue B, Smets B, Winderickx J, Accardo S, de Vos DE, Thevissen K (2006) Level of M(IP)2 C sphingolipid affects plant defensin sensitivity, oxidative stress resistance and chronological life-span in yeast. FEBS Lett 580:1903–1907
Baxter AA, Richter V, Lay FT, Poon IKH, Adda CG, Veneer PK, Phan TK, Bleackley MR, Anderson MA, Kvansakul M, Hulett MD (2015) The tomato defensin TPP3 binds phosphatidylinositol (4,5)-bisphosphate via a conserved dimeric cationic grip conformation to mediate cell lysis. Mol Cell Biol 35:1964–1978
Baul U, Kuroda K, Vemparala S (2014) Interaction of multiple biomimetic antimicrobial polymers with model bacterial membranes. J Chem Phys 141:084902
Bocchinfuso G, Palleschi A, Orioni B, Grande G, Formaggio F, Toniolo C, Park Y, Hahm K, Stella L (2009) Different mechanisms of action of antimicrobial peptides: insights from fluorescence spectroscopy experiments and molecular dynamics simulations. J Pept Sci 15:550–558
Bonucci A, Balducci E, Pistolesi S, Pogni R (2013) The defensin–lipid interaction: Insights on the binding states of the human antimicrobial peptide HNP-1 to model bacterial membranes. Biochim Biophys Acta 1828:758–764
Bulacu M, Sevink GJA (2015) Computational insight in the role of fusogenic lipopeptides at the onset of liposome fusion. Biochim Biophys Acta 1848: 848–858
Colavita I, Nigro E, Sarnataro D, Scudiero O, Granata V, Daniele A, Zagari A, Pessi A, Salvatore F (2015) Membrane protein 4F2/CD98 is a cell surface receptor involved in the internalization and trafficking of human β-defensin 3 in epithelial cells. Chem Biol 22:217–228
Cruz VL, Ramos J, Melo MN, Martinez SJ (2013) Bacteriocin AS-48 binding to model membranes and pore formation as revealed by coarse-grained simulations. Biochim Biophys Acta 1828: 2524–2531
de Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Scha LV, Periole X, Tieleman DP, Marrink SJ (2013) Improved parameters for the martini coarse-grained protein force field. J Chem Theory Comput 9:687–697
de Medeiros LN, Domitrovic T, de Andrade PC, Faria J, Bergter EB, Weissmüller G, Kurtenbach E (2014) Psd1 binding affinity toward fungal membrane components as assessed by SPR: the role of glucosylceramide in fungal recognition and entry. Biopolymers 102:456–464
Efimova SS, Malev VV, Ostroumova OS (2016) Effects of dipole potential modifiers on heterogenic lipid bilayers. J Membr Biol 249:97–106
Ermakova EA, Faizullin DA, Idiyatullin BZ, Khairutdinov BI, Mukhamedova LN, Tarasova NB, Toporkova YY, Osipova EV, Gogolev YV, Zuev YF, Nesmelova IV (2016) Structure of scots pine defensin 1 by spectroscopic methods and computational modeling. Int J Biological Macromol 84: 142–152
Flinner N, Schleiff E (2015) Dynamics of the glycophorin a dimer in membranes of native-like composition uncovered by coarse-grained molecular dynamics simulations. PLoS ONE 10:e0133999
Gurtovenko AA, Vattulainen I (2007) Molecular mechanism for lipid flip-flops. J Phys Chem B 111:13554–13559
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Hong C, Tieleman DP, Wang Y (2014) Microsecond molecular dynamics simulations of lipid mixing. Langmuir 30:11993–12001
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14.1: 33–38
Janosi L, Gorfe AA (2010) Simulating POPC and POPC/POPG bilayers: conserved packing and altered surface reactivity. J Chem Theory Comput 6:3267–3273
Jo S, Lim JB, Klauda JB, Im W (2009) CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J 97:50–58
Khairutdinov BI, Bessolicina EK, Efimov SV, Toporkova YY, Tarasova NB, Ermakova EA, Zuev YF (2016) Investigation of structural-dynamics properties of Scots pine defensin 1 by NMR spectroscopy. International symposium «Magnetic resonance: from fundamental research to practical applications». Book of abstracts, Kazan, p 186
Kovaleva V, Kiyamova R, Cramer R, Krynytskyy H, Gout I, Filonenko V, Gout R (2009) Purification and molecular cloning of antimicrobial peptides from Scots pine seedlings. Peptides 30:2136–2143
Kučerka N, Nieh MP, Katsaras J (2011) Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta 1808: 2761–2771
Kučerka N, Holland BW, Gray CC, Tomberli B, Katsaras J (2012) Scattering density profile model of POPG bilayers as determined by molecular dynamics simulations and small-angle neutron and x-ray scattering experiments. J Phys Chem B 116:232–239
Kvansakul M, Lay FT, Adda CG, Veneer PK, Baxter AA, Phan TK, Poon IKH, Hulett MD (2016) Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin–lipid oligomeric assemblies and membrane permeabilization. Proc Natl Acad Sci USA 113:11202–11207
Lay FT, Mills GD, Poon IK, Cowieson NP, Kirby N, Baxter AA, van der Weerden NL, Dogovski C, Perugini MA, Anderson MA, Kvansakul M, Hulett MD (2012) Dimerization of plant defensin NaD1 enhances its antifungal activity. J Biol Chem 287:19961–19972
Lee J, Jung SW, Cho AE (2016) Molecular insights into the adsorption mechanism of human β-defensin-3 on bacterial membranes. Langmuir 32:1782–1790
Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW (1996) Membrane pores induced by magainin. BioChemistry 35:13723–13728
Marrink SJ, de Vries AH, Tieleman DP (2009) Lipids on the move: simulations of membrane pores, domains, stalks and curves. Biochim Biophys Acta 1788:149–168
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111:7812–7824
Midura-Nowaczek K, Markowska A (2014) Antimicrobial peptides and their analogs: searching for new potential therapeutics. Perspect Medicin Chem 6:73–80
Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ (2008) The MARTINI coarse-grained force field: extension to proteins. J Chem Theory Comput 4:819–834
Periole X, Cavalli M, Marrink SJ, Ceruso MA (2009) Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J Chem Theory Comput 5:2531–2543
Poon IK, Baxter AA, Lay FT, Mills GD, Adda CG, Payne JA, Phan TK, Ryan GF, White JA, Veneer PK, van der Weerden NL, Anderson MA, Kvansakul M, Hulett MD (2014) Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. Elife 3:e01808
Qi Y, Ingólfsson HI, Cheng X, Lee J, Marrink SJ, Im W (2015) CHARMM-GUI martini maker for coarse-grained simulations with the martini force field. J Chem Theory Comput 11:4486–4494
Sagaram US, El-Mounadi K, Buchko GW, Berg HR, Kaur J, Pandurangi RS, Smith TJ, Shah DM (2013) Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: identification of an RGFRRR motif governing fungal cell entry. PLoS ONE 8:e82485
Sani MA, Separovic F (2016) How membrane-active peptides get into lipid membranes. Acc Chem Res 49:1130–1138
Sengupta D, Leontiadou H, Mark AE, Marrink SJ (2008) Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta 1778: 2308–2317
Skjevik AA, Madej BD, Dickson CJ, Teigen K, Walker RC, Gould IR (2015) All-atom lipid bilayer self-assembly with the AMBER and CHARMM lipid force fields. Chem Commun (Camb) 51:4402–4404
Shai Y, Oren Z (2001) From ‘‘carpet’’ mechanism to de novo designed diastereomeric cell-selective antimicrobial peptides. Peptides 22:1629–1641
Soblosky L, Ramamoorthy A, Chen Z (2015) Membrane interaction of antimicrobial peptides using E. coli lipid extract as model bacterial cell membranes and SFG spectroscopy. Chem Phys Lipids 187:20–33
Su Y, Li S, Hong M (2013) Cationic membrane peptides: atomic-level insight of structure–activity relationships from solid-state NMR. Amino Acids 44:821–833
Thevissen K, Osborn RW, Acland DP, Broekaert WF (1997) Specific, high affinity binding sites for an antifungal plant defensin on neurospora crassa hyphae and microsomal membranes. J Biol Chem 272:32176–32181
Thevissen K, de Mello Tavares P, Xu D, Blankenship J, Vandenbosch D, Idkowiak-Baldys J, Govaert G, Bink A, Rozental S, De Groot PW (2012) The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans. Mol Microbiol 84:166–180
van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible and free. J Comp Chem 26:1701–1719
van der Weerden NL, Hancock REW, Anderson MA (2010) Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J Biol Chem 285:37513–37520
van Eerden FJ, de Jong DH, de Vries AH, Wassenaar TA, Siewert J., Marrink SJ (2015) Characterization of thylakoid lipid membranes from cyanobacteria and higher plants by molecular dynamics simulations. Biochim Biophys Acta 1848: 1319–1330
Wang Y, Schlamadinger DE, Kim JE, McCammon JA (2012) Comparative molecular dynamics simulations of the antimicrobial peptide CM15 in model lipid bilayers. Biochim Biophys Acta 1818: 1402–1409
Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP (2014) Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J Chem Theory Comput 10:676–690
Wu Y, He K, Ludtke SJ, Huang HW (1995) X-ray diffraction study of lipid bilayer membranes interacting with amphiphilic helical peptides: diphytanoyl phosphatidylcholine with alamethicin at low concentrations. Biophys J 68:2361–2369
Zhao X, Yu H, Yang L, Li Q, Huang X (2015) Simulating the antimicrobial mechanism of human β-defensin-3 with coarse-grained molecular dynamics. J Biomol Struct Dyn 33:2522–2531
Acknowledgements
We thank Moscow State University for providing access to the SKIF-MGU ‘‘Lomonosov’’ supercomputer where all simulations were run. This work was supported by grants from the Russian Foundation for Basic Research and the Government of Tatarstan Republic (No. 15-44-02309) and from the Russian Foundation for Basic Research (No. 15-29-01239). Part of the work was performed in accordance with the Russian Government Program of Competitive Growth of Kazan Federal University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ermakova, E., Zuev, Y. Interaction of Scots Pine Defensin with Model Membrane by Coarse-Grained Molecular Dynamics. J Membrane Biol 250, 205–216 (2017). https://doi.org/10.1007/s00232-017-9950-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-017-9950-5