Abstract
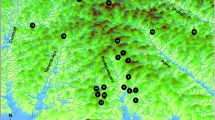
Genetic diversity and population structure of snapper (Pagrus auratus, Bloch and Schneider), a coastal demersal sparid fish, were determined using six nuclear microsatellite loci and SSCP (single strand conformational polymorphism) analysis of themitochondrial (mt) DNA D-loop in samples collected across the range of the species in New Zealand. Microsatellite data showed similar results to allozyme data collected in the late 1970s that found differentiation between the north-east and southern populations. In addition, an isolated population of snapper in Tasman Bay was identified. The two data sets provide evidence for the temporal stability of the genetic population structure of snapper over 22 years, with differentiation over relatively small spatial scales separated by oceanographic boundaries rather than isolation by distance. In contrast to nuclear markers, mtDNA did not reveal any significant genetic heterogeneity among samples.


Similar content being viewed by others
References
Adcock G, Ramirez B, Hauser L, Smith P, Carvalho G (2000) Screening of DNA polymorphisms in samples of archived scales from New Zealand snapper. J Fish Biol 56:1283–1287
Allegrucci GC, Fortunato C, Sbordoni V (1997) Genetic structure and allozyme variation of sea bass (Dicentrarchus labrax and D. punctatus) in the Mediterranean Sea. Mar Biol 128:347–358
Allendorf FW, Seeb LW (2000) Concordance of genetic divergence among sockeye salmon populations at allozyme, nuclear DNA, and mitochondrial DNA markers. Evolution 54:640–651
Annala JH, Sullivan KJ, O'Brien CJ, Iball SD (2000) Report from the Fisheries Assessment Plenary, May 1998: stock assessment and yield estimates. Available from the NIWA Library, Wellington, New Zealand
Bagley MJ, Lindquist DG, Geller JB (1999) Microsatellite variation, effective population size, and population genetic structure of vermillion snapper Rhomboplites aurorubens, off the southeastern USA. Mar Biol 134:609–620
Ball AO, Sedberry GR, Zatcoff MS, Chapman RW, Carlin JL (2000) Population structure of the wreckfish Polyprion americanus determined with microsatellite genetic markers. Mar Biol 137:1077–1090
Begg GA, Keenan CP, Sellin MJ (1998) Genetic variation and stock structure of school mackerel and spotted mackerel in northern Australian waters. J Fish Biol 53:543–559
Bembo DG, Carvalho GR, Cingolani N, Arneri E, Giannetti G, Pitcher TJ (1996) Allozymic and morphometric evidence for two stocks of the European anchovy Engraulis encrasicolus in Adriatic waters. Mar Biol 126:529–538
Birky CW, Fuerst P, Maruyama T (1989) Organelle gene diversity under migration, mutation and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparisons to nuclear genes. Genetics 121:613–627
Blackwell RG, Gilbert DJ, Davies NM (2000) Age composition of commercial snapper landings in SNA 2 and Tasman Bay/Golden Bay (SNA7), 1998–99. New Zealand Fisheries Assessment Report 2000/12, NIWA Library, Wellington, New Zealand
Brown BL, Epifanio JM, Smouse PE, Kobak CJ (1996) Temporal stability of mtDNA haplotype frequencies in American shad stocks: to pool or not to pool across years? Can J Fish Aquat Sci 53:2274–2283
Bruford MW, Hanotte O, Brookfield JF, Burke T (1992) Single locus and multilocus DNA fingerprinting. In: Hoelzel AR (ed) Molecular genetic analysis of populations—a molecular approach. IRL, Oxford, pp 227–229
Cassie RM (1956) Early development of the snapper, Chrysophrys auratus Forster. Trans R Soc NZ 83:705–713
Chiswell SM (2000) The Wairarapa coastal current. NZ J Mar Freshw Res 34:303–315
Crossland J (1982) Movements of tagged snapper in the Hauraki Gulf. Occasional publication no. 35, Fisheries Research Division, New Zealand Ministry of Agriculture and Fisheries, Wellington
Donnellan SC, McGlennon D (1994) Stock identification and discrimination in snapper (Pagrus auratus) in Southern Australia. Project report 94/168, Fisheries Research and Development Corporation, Henley Beach, Australia
Gauldie RW, Wood R (2002) The implications for marine reserves from the movement of tagged snapper Pagrus auratus in the Tasman Bay/Golden Bay fishery. Fish Oceanogr (in press)
Gilbert DJ, McKenzie JR (1999) Sources of bias in biomass estimates from tagging programmes in the SNA 1 snapper (Pagrus auratus) stock. New Zealand Fisheries Assessment Research Document 99/16, NIWA Library, Wellington, New Zealand
Gold JR, Richardson LR, Turner TF (1999) Temporal stability and spatial divergence of mitochondrial DNA haplotype frequencies in red drum (Sciaenops ocellatus) from coastal regions of the western Atlantic Ocean and Gulf of Mexico. Mar Biol 133:593–602
Goodman SJ (1997) RST-CALC: a collection of computer programs for calculating unbiased estimates of genetic differentiation and gene flow from microsatellite data and determining their significance. Mol Ecol 6:881–885
Goudet J (1995) F_STAT (1.2), a program for IBM compatible PCs to calculate Weir and Cockerman's (1984) estimators of F-statistics. J Hered 86:485–486
Hauser L, Ward RD (1998) Population identification of pelagic fish. In: Carvalho G (ed) Advances in molecular ecology. IOS, Amsterdam, pp 191–224
Hauser L, Adcock GJ, Smith PJ, Bernal Ramírez JH, Carvalho GR (2002) Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc Natl Acad Sci USA 99:11742–11747
Heath RA (1985) A review of physical oceanography of the seas around New Zealand—1982. NZ J Mar Freshw Res 19:79–124
Hedgecock D (1994) Does variance in reproductive success limit effective population size in marine organisms? In: Beaumont AR (ed) Genetics and evolution of aquatic organisms Chapman and Hall, London, pp 122–134
Johnson MS, Creagh S, Moran M (1986) Genetic subdivision of stocks of snapper Chryspophrys unicolor, in Shark Bay, Western Australia. Aust J Mar Freshw Res 37:337–345
Kotoulas G, Bonhomme F, Borsa P (1995) Genetic structure of the common sole Solea vulgaris at different geographic scales. Mar Biol 122:361–375
Larson RJ, Julian RM (1999) Spatial and temporal genetic patchiness in marine populations and their implications for fisheries management. Cal Coop Ocean Fish Investig Rep 40:94–99
Lemaire CG, Allegrucci G, Naciri M, Bahri-Sfar L, Kara H, Bonhomme F (2000) Do discrepancies between microsatellite and allozyme variation reveal differential selection between sea and lagoon in the sea bass (Dicentrarchus labrax)? Mol Ecol 9:457–467
Longhurst AR (1958) Racial differences in size and growth of snapper. NZ J Sci 1:487–499
Lundy CJ, Rico C, Hewitt GM (2000) Temporal and spatial genetic variation in spawning grounds of European hake (Merluccius merluccius) in the Bay of Biscay. Mol Ecol 9:2067–2079
MacDonald CM (1980) Population structure, biochemical adaptation and systematics in temperate marine fishes of the genus Arripis and Chrysophrys (Pisces: Perciformes). PhD thesis, Australian National University, Canberra
McElroy D, Moran P, Bermingham E, Kornfield I (1992) REAP: an integrated environment for the manipulation and phylogenetic analysis of restriction data. J Hered 83:157–158
Mork J, Ryman N, Stahl G, Utter F, Sundnes G (1985) Genetic variation in Atlantic cod (Gadus morhua) throughout its range. Can J Fish Aquat Sci 42:1580–1587
Orita M, Suzuki Y, Seiya T, Hayashi K (1989) A rapid and sensitive detection of point mutations and genetic polymorphisms using polymerase chain reaction. Genomics 5:874–879
Paul LJ (1967) An evaluation of tagging experiments on the New Zealand snapper Chrysophrys auratus (Forster), during the period 1952 to 1963. NZ J Mar Freshw Res 1:455–463
Paul LJ, Tarring SC (1980) Growth rate and population structure of snapper Chrysophrys auratus (Forster) in the East Cape region, New Zealand. NZ J Mar Freshw Res 14:237–247
Paulin CD (1990) Pagrus auratus, a new combination for the species known as "snapper" in Australasian waters (Pisces: Sparidae). NZ J Mar Freshw Res 24:259–265
Perez-Enriquez R, Taniguchi N (1999) Genetic structure of red sea bream (Pagrus major) population off Japan and the Southwest Pacific, using microsatellite DNA markers. Fish Sci (Tokyo) 65:23–30
Pogson GH, Mesa KA, Boutilier RG (1995) Genetic population-structure and gene flow in the Atlantic cod Gadus morhua—a comparison of allozyme and nuclear RFLP loci. Genetics 139:375–385
Pogson GH, Taggart CT, Mesa KA, Boutilier RG (2001) Isolation by distance in the Atlantic cod, Gadus morhua, at large and small geographic scales. Evolution 55:131–146
Purcell MK, Kornfield I, Fogarty M, Parker A (1996) Interdecadal heterogeneity in mitochondrial DNA of Atlantic haddock (Melanogrammus aeglefinus) from Georges Bank. Mol Mar Biol Biotechnol 5:185–192
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reynolds J, Weir BS, Cockerham CC (1983) Estimation of the co-ancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–779
Roff DA, Bentzen P (1989) The statistical analysis of mitochondrial DNA polymorphisms: χ2 and the problem of small samples. Mol Biol Evol 6:539–545
Ruzzante DE, Taggart CT, Doyle RW, Cook D (2001) Stability in the historical pattern of genetic structure of Newfoundland coast (Gadus morhua) despite the catastrophic decline in population size from 1964 to 1994. Conserv Genet 2:257–269
Sambrook E, Fritsch F, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Schneider S, Kueffer J-M, Roessli D, Excoffier L (1997) Arlequin ver. 1.1: a software programme for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva
Shaw PW, Turan C, Wright JM (1999) Microsatellite DNA analysis of population stucture in Atlantic herring (Clupea harengus), with direct comparison to allozyme and mtDNA RFLP analyses. Heredity 83:490–499
Sokal RR, Rohlf FJ (1995) Biometry. Freeman, New York
Smith PJ (1979) Esterase gene frequencies and temperature relationships in the New Zealand snapper Chrysophrys auratus. Mar Biol 53:305–310
Smith PJ, Francis RICC (1983) Relationship between esterase gene frequencies and length in juvenile snapper Chrysophrys auratus Forster. Anim Blood Groups Biochem Genet 14:151–158
Smith PJ, Francis RICC, Paul LJ (1978) Genetic variation and population structure in the New Zealand snapper. NZ J Mar Freshw Res 12:343–350
Tabata K, Mizuta A (1997) RFLP analysis of the mtDNA D-loop region in red sea bream Pagrus major population from four locations of western Japan. Fish Sci (Tokyo) 63:211–217
Tabata K, Taniguchi N (2000) Differences between Pagrus major and Pagrus auratus through mainly mtDNA control region analysis. Fish Sci (Tokyo) 66:9–18
Taniguchi N, Sugama K (1990) Genetic variation and population structure of red sea bream Pagrus major in the coastal waters of Japan and the East China Sea. Nippon Suisan Gakk 56:1069–1077
Takagi M, Taniguchi N, Cook D, Doyle RW (1997) Isolation and characterization if microsatellite loci from red sea bream (Pagrus major) and detection in closely related species. Fish Sci (Tokyo) 63:199–204
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. Heredity 89:438–450
Ward RD, Woodwark M, Skibinski DOF (1994) A comparison of genetic diversity levels in marine, freshwater and anadromous fishes. J Fish Biol 44:213–232
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Willis TJ, Parsons DM, Babcock RC (2001) Evidence for long-term site fidelity of snapper (Pagrus auratus) within a marine reserve. NZ J Mar Freshw Res 35:581–590
Zeldis JR, Francis RICC (1998) A daily egg production method estimate of snapper biomass in Hauraki Gulf, New Zealand. ICES J Mar Sci 55:522–534
Acknowledgements
We thank R. Blackwell, E. Bowman, H. Cadenhead, J. Holdsworth, D. Parkinson and M. Smith for help with sample collection. The laboratory work was supported by the Leverhulme Trust (grant reference F/181/Q).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M.S. Johnson, Crawley
Rights and permissions
About this article
Cite this article
Bernal-Ramírez, J.H., Adcock, G.J., Hauser, L. et al. Temporal stability of genetic population structure in the New Zealand snapper, Pagrus auratus, and relationship to coastal currents. Marine Biology 142, 567–574 (2003). https://doi.org/10.1007/s00227-002-0972-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-002-0972-9