Abstract
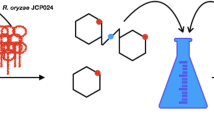
The microbial metabolic activity during malting is an important factor affecting malt quality besides barley varieties. Elucidating the effect and action mechanism of dominant microbial on malt quality will provide a new strategy for the precise control of malt quality. Pantoea spp. was identified as the dominant bacteria of barley by the aid of high-throughput sequencing technique. Moreover, the dilution plating results showed that P. agglomerans was the dominant bacteria in quantity. During malting, the number of P. agglomerans did not change significantly after steeping, but increased exponentially during germination. The quality of malt, especially the filtration rate, is affected by the inoculation of P. agglomerans in the malting process. The exopolysaccharides of P. agglomerans was the main factor leading to the decrease of wort filtration speed. When the exopolysaccharides of P. agglomerans in the wort reached at 12.5 μg/mL, the filtration speed decreased by 50%.




Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Nischwitz R, Cole NW, MacLeod L (1999) Malting for brewhouse performance. J Inst Brew 105:219–227
Van Nierop SNE, Rautenbach M, Axcell BC, Cantrell IC (2018) The impact of microorganisms on barley and malt quality-A review. J Am Soc Brew Chem 64:69–78
Mastanjevic K, Krstanovic V, Mastanjevic K, Sarkanj B (2018) Malting and brewing industries encounter fusarium spp related problems. Fermentation 4:15
Virkajärvi V, Sarlin T, Laitila A (2018) Fusarium profiling and barley malt gushing propensity. J Am Soc Brew Chem 75:181–192
Geissinger C, Whitehead I, Hofer K, Hess M, Habler K, Becker T, Gastl M (2019) Influence of Fusarium avenaceum infections on barley malt: monitoring changes in the albumin fraction of barley during the malting process. Int J Food Microbiol 293:7–16
Bianco A, Fancello F, Balmas V, Zara G, Dettori M, Budroni M (2018) The microbiome of Sardinian barley and malt. J Inst Brew 124:344–351
Bokulich N, Bamforth C (2013) The microbiology of malting and brewing. Microbiol Mol Biol Rev 77:157–172
Lowe DP, Arendt EK (2004) The use and effects of lactic acid bacteria in malting and brewing with their relationships to antifungal activity, mycotoxins and gushing: a review. J Inst Brew 110:163–180
Briggs DE (2002) Malt modification - a century of evolving views. J Inst Brew 108:395–405
Luziatelli F, Ficca AG, Bonini P, Muleo R, Gatti L, Meneghini M, Tronati M, Melini F, Ruzzi M (2020) A genetic and metabolomic perspective on the production of indole-3-acetic acid by P agglomerans and use of their metabolites as biostimulants in plant nurseries. Front Microbiol 11:1475
Greiner R (2004) Degradation of myo-inositol hexakisphosphate by a phytate-degrading enzyme from Pantoea agglomerans. Protein J 23:577–585
Wang J, Shao Z, Hong Y, Li C, Liu FZ (2010) A novel β-mannanase from P. agglomerans A021: gene cloning, expression, purification and characterization. World J Microb Biot 26:1777–1784
Guo L, Luo Y, Zhou Y, Bianba C, Guo H, Zhao Y, Fu H (2020) Exploring microbial dynamics associated with flavours production during highland barley wine fermentation. Food Res Int 130:108971
Qin Q, Liu J, Hu S, Yu J, Zhang L, Huang S, Yang M, Wang L (2022) Exploring Fungal Species Diversity in the premature yeast flocculation (PYF) of barley malt using deep sequencing. J Am Soc Brew Chem. https://doi.org/10.1080/03610470.2021.2025329
European Brewery Convention Analysis Committee (2006) Analytica-EBC. Fachverlag Hans Carl, Nurnberg
Geissinger C, Gastl M, Becker T (2022) Enzymes from cereal and Fusarium metabolism involved in the malting process - A review. J Am Soc Brew Chem 80:1–16
Gastl M, Kupetz M, Becker T (2020) Determination of Cytolytic Malt Modification – Part II: Impact on wort separation. J Am Soc Brew Chem 79:66–74
Kanjan P, Sahasrabudhe NM, de Haan BJ, de Vos P (2017) Immune effects of β-glucan are determined by combined effects on Dectin-1, TLR2, 4 and 5. J Funct Foods 37:433–440
Ostlie HM, Porcellato D, Kvam G, Wicklund T (2021) Investigation of the microbiota associated with ungerminated and germinated Norwegian barley cultivars with focus on lactic acid bacteria. Int J Food Microbiol 341:109059
Li X, Cai G, Wu D, Zhang M, Lin C, Lu J (2018) Microbial community dynamics of Dan’er barley grain during the industrial malting process. Food Microbiol 76:110–116
Normander B, Prosser JI (2000) Bacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions. Appl Environ Microbiol 66:4372–4377
Wright SA, Zumoff CH, Schneider L, Beer SV (2001) P. agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl Environ Microbiol 67:284–292
Slaughter JC (1979) Production of wort from unmalted barley using bacterial enzymes. IFST Proceedings 12:171–174
Dutkiewicz J, Mackiewicz B, Lemieszek MK, Golec M, Milanowski J (2015) Pantoea agglomerans: a marvelous bacterium of evil and good.part I. deleterious effects: Dust-borne endotoxins and allergens - focus on cotton dust. Ann Agric Environ Med 22:576–588
Yang L, Schröder P, Vestergaard G, Schloter M, Radl V (2020) Response of barley plants to drought might be associated with the recruiting of soil-borne endophytes. Microorganisms 8:1414
Felsociova S, Kowalczewski PL, Krajcovic T, Drab S, Kacaniova M (2020) Quantitative and qualitative composition of bacterial communities of malting barley grain and malt during long-term storage. Agronomy-Basel 10:15
Laitila A, Kotaviita PP, Home S, Wilhelmson A (2007) Indigenous microbial community of barley greatly influences grain germination and malt quality. J Inst Brew 113:9–20
Booysen C, Dicks LMT, Meijering I, Ackermann A (2002) Isolation, identification and changes in the composition of lactic acid bacteria during the malting of two different barley cultivars. Int J Food Microbiol 76:63–73
Mangan D, Cornaggia C, Liadova A, Draga A, Ivory R, Evans DE, McCleary BV (2018) Development of an automatable method for the measurement of endo-1,4-β-xylanase activity in barley malt and initial investigation into the relationship between endo-1,4-β-xylanase activity and wort viscosity. J Cereal Sci 84:90–94
Baek EJ, Kwon YA, Hong KW (2016) Adding enzymes to improve the properties of the Korean barley Gwangmaek wort during mashing. Food Sci Biotechnol 25:1387–1391
Laitila A, Manninen J, Priha O, Smart K, Tsitko I, James S (2018) Characterisation of barley-associated bacteria and their impact on wort separation performance. J Inst Brew 124:314–324
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31871785), and the Program of Introducing Talents of Discipline to Universities (111 Project) (111-2-06)
Author information
Authors and Affiliations
Contributions
Conceptualization: BH and GC; methodology: BH, YX, MZ, and GC; formal analysis and investigation: BH and GC; writing—original draft preparation: BH; writing—review and editing: BH and GC; funding acquisition: GC; supervision: GC and JL.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have not known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Human and animal research
This work does not involve any human or animal testing.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, B., Xie, Y., Zhang, M. et al. Impact of barley endophytic Pantoea agglomerans on the malt filterability. Eur Food Res Technol 249, 1403–1409 (2023). https://doi.org/10.1007/s00217-023-04223-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-023-04223-y