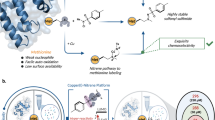
Abstract
Advances in high-throughput high-resolution mass spectrometry and the development of thermal proteome profiling approach (TPP) have made it possible to accelerate a drug target search. Since its introduction in 2014, TPP quickly became a method of choice in chemical proteomics for identifying drug-to-protein interactions on a proteome-wide scale and mapping the pathways of these interactions, thus further elucidating the unknown mechanisms of action of a drug under study. However, the current TPP implementations based on tandem mass spectrometry (MS/MS), associated with employing lengthy peptide separation protocols and expensive labeling techniques for sample multiplexing, limit the scaling of this approach for the ever growing variety of drug-to-proteomes. A variety of ultrafast proteomics methods have been developed in the last couple of years. Among them, DirectMS1 provides MS/MS-free quantitative proteome-wide analysis in 5-min time scale, thus opening the way for sample-hungry applications, such as TPP. In this work, we demonstrate the first implementation of the TPP approach using the ultrafast proteome-wide analysis based on DirectMS1. Using a drug topotecan, which is a known topoisomerase I (TOP1) inhibitor, the feasibility of the method for identifying drug targets at the whole proteome level was demonstrated for an ovarian cancer cell line.
Graphical Abstract





Similar content being viewed by others
References
Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–55. https://doi.org/10.1038/nature19949.
Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. https://doi.org/10.1038/msb.2011.82.
Saei AA, Sabatier P, Tokat ÜG, Chernobrovkin A, Pirmoradian M, Zubarev RA. Comparative proteomics of dying and surviving cancer cells improves the identification of drug targets and sheds light on cell life/death decisions. Mol Cell Proteomics MCP. 2018;17:1144–55. https://doi.org/10.1074/mcp.RA118.000610.
Gaetani M, Sabatier P, Saei AA, Beusch CM, Yang Z, Lundström SL, et al. Proteome integral solubility alteration: a high-throughput proteomics assay for target deconvolution. J Proteome Res. 2019;18:4027–37. https://doi.org/10.1021/acs.jproteome.9b00500.
Savitski MM, Reinhard FBM, Franken H, Werner T, Savitski MF, Eberhard D, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346:1255784. https://doi.org/10.1126/science.1255784.
Mateus A, Kurzawa N, Perrin J, Bergamini G, Savitski MM. Drug target identification in tissues by thermal proteome profiling. Annu Rev Pharmacol Toxicol. 2022;62:465–82. https://doi.org/10.1146/annurev-pharmtox-052120-013205.
Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–7. https://doi.org/10.1126/science.1233606.
Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M. High-resolution enabled TMT 8-plexing. Anal Chem. 2012;84:7188–94. https://doi.org/10.1021/ac301553x.
Werner T, Sweetman G, Savitski MF, Mathieson T, Bantscheff M, Savitski MM. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal Chem. 2014;86:3594–601. https://doi.org/10.1021/ac500140s.
Sauer P, Bantscheff M. Thermal proteome profiling for drug target identification and probing of protein states. In: Gevaert, K. (eds) Mass spectrometry-based proteomics. Methods in Molecular Biology. 2023. https://doi.org/10.1007/978-1-0716-3457-8_5.
George AL, Sidgwick FR, Watt JE, Martin MP, Trost M, Marín-Rubio JL, et al. Comparison of quantitative mass spectrometric methods for drug target identification by thermal proteome profiling. J Proteome Res. 2023. https://doi.org/10.1021/acs.jproteome.3c00111.
Ivanov MV, Bubis JA, Gorshkov V, Abdrakhimov DA, Kjeldsen F, Gorshkov MV. Boosting MS1-only proteomics with machine learning allows 2000 protein identifications in single-shot human proteome analysis using 5 min HPLC Gradient. J Proteome Res. 2021;20:1864–73. https://doi.org/10.1021/acs.jproteome.0c00863.
Ivanov MV, Bubis JA, Gorshkov V, Tarasova IA, Levitsky LI, Solovyeva EM, et al. DirectMS1Quant: ultrafast quantitative proteomics with MS/MS-free mass spectrometry. Anal Chem. 2022;94:13068–75. https://doi.org/10.1021/acs.analchem.2c02255.
Zhou Y, Zhao HY, Jiang D, Wang LY, Xiang C, Wen SP, et al. Low toxic and high soluble camptothecin derivative 2–47 effectively induces apoptosis of tumor cells in vitro. Biochem Biophys Res Commun. 2016;472:477–81. https://doi.org/10.1016/j.bbrc.2016.02.015.
Kazakova EM, Solovyeva EM, Levitsky LI, Bubis JA, Emekeeva DD, Antonets AA, et al. Proteomics-based scoring of cellular response to stimuli for improved characterization of signaling pathway activity. Proteomics. 2023;23:e2200275. https://doi.org/10.1002/pmic.202200275.
Hulstaert N, Shofstahl J, Sachsenberg T, Walzer M, Barsnes H, Martens L, et al. ThermoRawFileParser: modular, scalable, and cross-platform RAW file conversion. J Proteome Res. 2020;19:537–42. https://doi.org/10.1021/acs.jproteome.9b00328.
Levitsky LI, Ivanov MV, Lobas AA, Gorshkov MV. Unbiased false discovery rate estimation for shotgun proteomics based on the target-decoy approach. J Proteome Res. 2017;16:393–7. https://doi.org/10.1021/acs.jproteome.6b00144.
Franken H, Mathieson T, Childs D, Sweetman GMA, Werner T, Tögel I, et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc. 2015;10:1567–93. https://doi.org/10.1038/nprot.2015.101.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. https://doi.org/10.1038/75556.
Pourquier P, Lansiaux A. Molecular determinants of response to topoisomerase I inhibitors. Bull Cancer (Paris). 2011;98:1287–98. https://doi.org/10.1684/bdc.2011.1474.
Thomas A, Pommier Y. Topoisomerase I in the Era of Precision Medicine. Clin Cancer Res. 2019;25:6581–9. https://doi.org/10.1158/1078-0432.CCR-19-1089.
Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2001;41:53–77. https://doi.org/10.1146/annurev.pharmtox.41.1.53.
Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci USA. 2002;99(24):15387–92. https://doi.org/10.1073/pnas.242259599.
Hertzberg RP, Caranfa MJ, Hecht SM. On the mechanism of topoisomerase I inhibition by camptothecin: evidence for binding to an enzyme-DNA complex. Biochemistry. 1989;28(11):4629–38. https://doi.org/10.1021/bi00437a018.
Trask DK, Muller MT. Stabilization of type I topoisomerase-DNA covalent complexes by actinomycin D. Proc Natl Acad Sci USA. 1988;85(5):1417–21. https://doi.org/10.1073/pnas.85.5.1417.
Funding
This work was supported by Russian Science Foundation (grant № 20–14-00229-P).
Author information
Authors and Affiliations
Contributions
Ivan I. Fedorov: data curation; formal analysis; visualization; writing, original draft preparation; writing, review and editing. Julia A. Bubis: methodology, investigation, laboratory work. Elizaveta M. Kazakova: investigation, laboratory work, data curation. Anna A. Lobas: methodology. Mark V. Ivanov: software, resources. Daria D. Emekeeva: investigation, laboratory work. Irina A. Tarasova: resources, formal analysis, supervision. Alexey A. Nazarov: investigation, laboratory work. Mikhail V. Gorshkov: conceptualization, supervision, funding acquisition, project administration, writing—review and editing. All authors discussed the results and commented on the manuscript. All authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fedorov, I.I., Bubis, J.A., Kazakova, E.M. et al. On the utility of ultrafast MS1-only proteomics in drug target discovery studies based on thermal proteome profiling method. Anal Bioanal Chem (2024). https://doi.org/10.1007/s00216-024-05330-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00216-024-05330-9