Abstract
The past decades have marked the rise of metabolomics and lipidomics as the -omics sciences which reflect the most phenotypes in living systems. Mass spectrometry–based approaches are acknowledged for both quantification and identification of molecular signatures, the latter relying primarily on fragmentation spectra interpretation. However, the high structural diversity of biological small molecules poses a considerable challenge in compound annotation. Feature-based molecular networking (FBMN) combined with database searches currently sets the gold standard for annotation of large datasets. Nevertheless, FBMN is usually based on collision-induced dissociation (CID) data, which may lead to unsatisfying information. The use of alternative fragmentation methods, such as electron-activated dissociation (EAD), is undergoing a re-evaluation for the annotation of small molecules, as it gives access to additional fragmentation routes. In this study, we apply the performances of data-dependent acquisition mass spectrometry (DDA-MS) under CID and EAD fragmentation along with FBMN construction, to perform extensive compound annotation in the crude extracts of the freshwater sentinel organism Gammarus fossarum. We discuss the analytical aspects of the use of the two fragmentation modes, perform a general comparison of the information delivered, and compare the CID and EAD fragmentation pathways for specific classes of compounds, including previously unstudied species. In addition, we discuss the potential use of FBMN constructed with EAD fragmentation spectra to improve lipid annotation, compared to the classic CID-based networks. Our approach has enabled higher confidence annotations and finer structure characterization of 823 features, including both metabolites and lipids detected in G. fossarum extracts.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The advent of -omics sciences has represented a real revolution in many fields of biological research [1]. Among these, lipidomics and metabolomics investigate lipids and metabolites, respectively, defined as biological compounds with a molecular weight lower than 1500 Da found in cells, biological fluids, or tissues [2, 3]. Although these two fields have lagged behind other -omics approaches for a long time, the past decades have marked their expansion due to their close connection with the organism’s phenotype [4]. As the main analytical goal is to identify and/or quantify as many compounds as possible within an investigated system, powerful analytical techniques and interpretation tools are required. The use of a single analytical platform fails in exhaustive detection, identification, and quantification of compounds in a given sample, simultaneously. Mass spectrometry (MS) provides an efficient and sensitive platform for the analysis of biological matrices, even for reduced volumes of samples [5, 6]. MS-based targeted approaches focus on analyzing one or a few compounds within a complex matrix, offering higher sensitivity and specificity for quantification but at the expense of reduced metabolite coverage [7]. On the other hand, non-targeted approaches aim to measure all compounds present in a sample, providing only semi-quantitative analysis [8]. Non-targeted approaches performed on high-resolution instruments (FWMH 20.000–50.000) based on time-of-flight (TOF) analyzers and ultra-high resolution instruments based on Orbitrap and Fourier transform ion cyclotron resonance (FTICR) analyzers (with FWMH ranging from 50.000 up to 1∙106 depending on the operational mode, as hyphenation with chromatography or direct injection analysis) enable exact mass measurements with an accuracy ranging from 5 to 0.1 ppm, representing nowadays, the gold standard for compound discovery and identification [9]. In particular, metabolite and lipid annotation can be reached through the acquisition of tandem MS (MS/MS or MS2) spectra in which selected precursor ions are broken into fragments which nature depends on the analyte chemical structure [10]. As in metabolomics and lipidomics biological samples contain hundreds to thousands of compounds of different natures and structures, different MS analytical strategies have been developed to reduce sample complexity and acquire a large amount of MS/MS spectra at once. Among these, data-dependent acquisition (DDA-MS) and data-independent acquisition (DIA-MS) are the most known [11,12,13]. While DIA-MS has been developed principally for proteome studies, its application in lipidomics and metabolomics remains to be optimized, and robust and universal deconvolution algorithms for fragmentation spectra reconstruction are still required [14,15,16,17].
Nowadays, compound annotation is preferentially based on the use of DDA-MS and on database search or manual attribution of the fragmentation spectra [18]. However, one of the major bottlenecks is represented by the extensive number of unknown compounds that have not yet been discovered (the unknown-unknown) and, from another point of view, by incomplete or missing reference spectra for comparison (the known-unknown) [19,20,21].
Molecular networking (MN) has emerged as an efficient tandem data organization approach which allows finding mass spectra similarities (fragments and neutral losses) among all the fragmentation spectra and gathering in one same molecular family, compounds which present structural similarities, as they belong, for example, to a same molecular class [22,23,24,25]. In the presence of matched compounds, annotation of ions in the same family can be easily performed according to the molecular formula, exact mass difference from the matched compounds, similarity score (expressed as a cosine), and comparison of their fragmentation pathways. More recently, the t-distributed Stochastic Neighbor Embedding (t-SNE), an unsupervised non-linear dimensionality reduction technique, has been integrated into the annotation workflow as a complement to MN visualization [26, 27]. This alternative data analysis method accounts for MN mis-linked compounds which eventually possess structure similarities but different fragmentation pathways, by approaching them to the related family. MN construction which is strongly supported by GNPS and the MetGem platforms has notoriously been applied in the field of natural substance and drug discovery [25] and more recently to highlight specialized pathways and obtain atlases of involved molecules in biological and clinical research [28, 29]. Despite the use of MN becoming a routine practice, it is worth noting that annotation performances based on the obtained molecular families depend strictly on the amount of information contained in the fragmentation spectra. It means that, in the case of intact precursors or in the presence of a low number of fragments, the construction of MN may suffer from poor specificity or not lead to appropriate structure assignments.
Since the rise of soft ionization techniques, biomolecule structural analysis has been preferentially performed under collision-induced dissociation (CID). In CID, kinetically excited ions collide with a neutral gas and gradually acquire sufficient internal energy to cause fragmentation [30]. The process is controlled by a thermodynamic factor with energies classically in the range of 1–100 eV [31]. Although the databases are being enriched with new reference spectra, this abundance can hardly satisfy an exhaustive annotation of compounds, given the great structural diversity of existing metabolites and lipids.
To achieve more effective annotation of compounds, alternative strategies have been tested during the years, such as the use of additional intrinsic descriptors like collision cross-sectional area (CCS) [32, 33], chemical derivatization prior to fragmentation [34], ionization in form of Li, Cu, Co, Ag (and so on), adducts [35,36,37], or alternative dissociation methods as photon or electron-activated dissociation [38]. Moreover, it is worth mentioning that compound annotation, especially in the presence of complex mixtures containing many isobars and isomers (as for complex lipids samples), might strongly be improved by hyphenation with peculiar chromatography techniques. In addition to the classic reversed phase (RP) and hydrophilic interaction (HILIC) LC, which both are nowadays used for metabolite and lipid separation [39, 40], additional techniques such as supercritical fluid chromatography (SFC) have shown particular interest in coupling with MS analysis [41, 42].
In 1979, the pioneering work of Cody and Freiser [43, 44] has led to the advent of a new fragmentation mode for small singly charged organics, firstly implemented in FTICR instruments and referred to in the literature as electron impact excitation of ions from organics (EIEIO), electron-induced dissociation (EID), or electron-activated dissociation (EAD). For further discussion, we will employ the term EAD. In EAD, the dissociation of singly charged ions is triggered by the impact with free electrons at kinetic energy in the range of ~ 10–20 eV. In this case, dissociation can be induced either by relaxation from the electronic excited state to a high vibrational state, as in CID; or by homolytic cleavage caused by the formation of an exited radical state [45]. EAD has proven to be complementary to CID in compound annotation for small molecules to happen, up to reach near-complete structural characterization for peculiar lipid classes such as phosphatidylcholines (PC) [46], sphingomyelins (SM) [47], triacylglycerols (TG) [48], and eicosanoids [49]. In fact, EAD allows elucidation of the lipid’s head group identity, chain length, and localization of double bonds and provides discrimination of trans/cis stereoisomers [50]. EAD fragmentation showed high potential for structural elucidation producing a general increase of produced fragments, especially for sodium and potassium adducts [51,52,53]. However, most of these studies have been performed on pure standards and hand-on utilization of EAD on crude extracts by LC–MS has been deployed mostly for quantitative scopes or for annotation of glucuronide and sulfate derivates of the studied standards. To the best of our knowledge, there are no studies on the use of EAD fragmentation to perform comprehensive compound annotation on crude complex samples containing thousands of undisclosed molecules.
In this work, we apply and compare two non-targeted mass spectrometry approaches based on DDA-MS under CID and EAD fragmentation (called herein respectively as DDA-CID-MS and DDA-EAD-MS), followed by molecular networking construction to perform extensive compound annotation and wide characterization on a sentinel organism commonly used in ecotoxicology studies and environmental monitoring: the freshwater crustacean amphipod Gammarus fossarum [54].
G. fossarum organisms represent an important amount of the biomass in European lotic freshwater environments providing prey for secondary consumers (e.g., fish) and intervening in the food web of river ecosystems. Their use as bioindicator species of water quality has been justified because of their high sensitivity to diverse micropollutants as pharmaceuticals, metals, and pesticides and of their high rate of bioaccumulation of toxic compounds [55]. In the last 20 years, research on G. fossarum has been pushed to study the biological perturbations caused by environmental contaminants. Mass spectrometry–based proteomics and proteogenomics have contributed to fostering the knowledge of the molecular physiology of reproduction in G. fossarum and to investigating the mechanisms of action of some contaminants, such as pesticides or metal ions [56,57,58,59,60,61,62,63,64]. More recently, shotgun lipidomics or mass spectrometry imaging have been integrated to gain extensive knowledge on G. fossarum lipid composition and on their spatial distribution in different organ sections [65, 66]. These studies have led to the discovery of an unexpected class of sulfur-containing lipids and highlighted the presence of different isobaric and isomeric lipids with peculiar tissue distribution. However, except for sporadic cases [67], information on fine lipid structures and on the nature of most metabolites in G. fossarum is still missing. For this and as in the first step, a classical approach based on DDA-CID-MS spectra collection performed on methanol/water extracts obtained from two reproductive stages (C1 and D1) of female specimens has been performed. This approach enabled us to compile a list containing 823 features, which were annotated through MN, t-SNE construction, database search, and manual interpretation. The same samples, when analyzed by DDA-EAD-MS, then allowed us to evaluate the performance of an alternative fragmentation technique for compound annotation in a complex sample containing both metabolites and lipids. Specifically, we conducted a critical evaluation of the use of EAD fragmentation for compound annotation by comparing the number of fragments formed in the two fragmentation modes, the fragmentation mechanisms for case compounds belonging to each molecular family, and the information provided by constructing molecular networking in the two fragmentation modes.
Material and methods
Gammarid collection and maintenance
Gammarus fossarum organisms were collected in a watercress site near the Pollon River (45° 57′ 25.8″ N 5° 15′ 43.6″ E) in France from a source population routinely used in the laboratory of INRAE. Organisms were collected by kick sampling using a net and selected in the field according to their size by using a series of sieves (± 1 cm in length). Organisms were quickly transported to the laboratory and kept in 30 L tanks supplied with continuous drilled groundwater under constant aeration. A 16/8 h light/dark photoperiod was maintained. The temperature was kept at 16 ± 1 °C. Organisms were fed ad libitum with alder leaves (Alnus glutinosa). Freeze-dried worms (Tubifex tubifex) were provided as a dietary supplement twice a week. For this study, female gammarids were selected to perform further investigation.[60]. Females at the C1 and D1 molting stages were sampled according to specific phenotypic criteria [63]. These two stages represent two contrasted conditions of females regarding their reproductive cycle due to the synchronization of ovary development and molting dynamics in amphipods. During the inter-molting stage C1, females are in a previtellogenic status regarding the gonadic development, while they are in a vitellogenic status during the pre-molting stage D1. All gammarids were washed with deionized water, weighted, and individually flash-frozen in liquid nitrogen until further analysis.
Chemical and sample preparation
Metabolite extraction was performed according to a slightly modified multi-omics sample extraction for G. fossarum by Faugere et al. [68]. Entire frozen female gammarids (n = 6 for the C1 stage and n = 6 for the D1 stage) were individually homogenized in 2 mL tubes prefilled with beads (Precellys® CK28 Lysing Kit, Bertin) using 300 µL of a methanol/water mixture (80/20 v/v) on a homogenizer (Precellys® Evolution Touch, Bertin) at 5500 rpm during two cycles of 25 s each spaced by a 10 s interval. Samples were then centrifugated at 10.000 rpm for 15 min in order to separate the metabolite fraction from the insoluble residues and 250 µL of the supernatant was collected and treated with 200 µL of cold methanol in order to inactivate residual enzymes, vortexed and mixed during 20 min at 4 °C (Eppendorf ThermoMixer® C). Samples were centrifugated for 10 min and 400 µL of the obtained supernatant were evaporated to dryness under nitrogen. Each residue was resolubilized by adding 15 µL water/methanol (90:10, v/v). The six fractions per stage were then vortexed and pooled to obtain a unique technical replicate for the C1 and a unique technical replicate for the D1 stage (90 µL each, concentration factor ∼17.0). Reagent blanks were prepared accordingly and in the absence of gammarid organisms. Samples were subjected to mass spectrometry analysis immediately after preparation, in order to limit compound degradation. Water, acetonitrile, methanol, and formic acid used for ultra-high performance liquid chromatography and for the preparation of the samples were LC–MS grade (99% purity).
Data-dependent acquisition under collision-induced dissociation and electron-activated dissociation
For high-throughput metabolite annotation and structural characterization, G. fossarum samples (C1 and D1) were submitted to data-dependent acquisition (DDA-MS) in both positive and negative ionization mode, either under CID or EAD on a high-resolution ZenoTOF 7600 mass spectrometer (Sciex) [69] equipped with a ExionLC™ (Sciex) UHPLC system. CID and EAD conditions were kept separate and applied to various several sample injections. While it was technically feasible to perform both fragmentation modes for a single sample in a single injection, doing so would have necessitated a reduction in MS/MS event accumulation time to maintain an acceptable scan time.
For instance, when working with Top10 DDA-MS, employing an accumulation time of 0.1 s for a full scan (MS1) event and 0.1 s for the MS/MS events resulted in a total scan time of 1.238 s. This allowed us to acquire a minimum of 13–15 data points for each chromatographic peak. Integrating EAD fragmentation events within the same injection would have required (1) decreasing accumulation times for both MS1 and MS/MS events in CID and EAD to maintain the total scan time, risking compromised chromatographic definition and MS/MS data quality, particularly in EAD fragmentation mode; (2) reducing the number of selected and fragmented TopX precursor ions, limiting the interpretable MS/MS data. For this study, we opted to maintain a Top10 analysis and conducted CID and EAD fragmentation in separate injections. Top10 DDA-MS was performed without utilizing an inclusion list to carry out analyses in a single batch, ensuring sample integrity and maintaining consistent chromatographic conditions. Furthermore, an unbiased MS/MS spectra collection in both CID and EAD conditions was chosen over the use of an inclusion list, aligning with the exploratory nature of this study. It is noteworthy that DDA-MS operated under a “dynamic background subtraction” on the Sciex platform, ensuring minimal fragmentation of background signals.
Finally, it is essential to mention that while a CE spread was employed for DDA-CID-MS (where multiple spectra are acquired at stepped CE values across an energy range, and a final mean MS/MS spectrum is obtained), a fixed value of KE was used for DDA-EAD-MS. Except for the electron transmission coefficient (ETC) parameter, which allows varying the number of irradiating electrons according to the analyte charge state [45], it is currently not possible to set dynamic KE or a KE spread in a single run. The decision to maintain a CE spread for DDA-CID-MS was driven by the diverse nature of compounds present in G. fossarum samples, aiming for high-quality and informative CID MS/MS spectra. Each sample and the blank were analyzed in triplicates for each analytical mode. The chromatographic separation was carried out on a Kinetex® 2.6 µm F5 100 Å column (150 × 2.1 µm) from Phenomenex. This column, which is constituted by pentafluorophenyl propyl (PFP) core–shell phase, was chosen to retain compounds at a wide range of polarity and efficiently resolve isomeric species, and therefore as a compromise for the analysis of both small metabolites and lipids with mid-polar nature. The decision to utilize a single column for analyzing a fraction containing both metabolites and polar lipids was made with the intention of enhancing molecular networking construction and minimizing potential redundancy that could arise from analyzing separate fractions under different conditions.
The used liquid chromatography and mass spectrometry experimental parameters are resumed in Table S1.
Data processing
Raw.wiff files were converted to the mzXML centroided format using the peak peaking (Vendor algorithm) in MSConvert (version 3.0.23024-30a908d). Data were then transferred to MzMine2 [70,71,72] (version 2.53), which results in being very stable among the various versions of MzMine2. For the positive ion mode, processing was performed by splitting the DDA-MS analysis into two chromatographic periods in order to optimize the processing parameters for metabolites and lipids, which presented a remarkable difference in peak intensity. Parameters used for data pre-treatment are gathered in Table S2. After all modules were compiled and alignment, features present in blanks and in less than two analytical replicates for each reproductive stage were removed. Data processing was performed separately for the DDA-CID-MS and DDA-EAD-MS and the obtained feature lists were compared in order to keep features which matched both exact mass and tR in the analysis under the two fragmentation modes.
Metabolite annotation and parameters for molecular networking construction
Obtained feature lists were exported and then uploaded on MetGem (version 1.3.6) for molecular networking and t-SNE construction. For DDA-CID-MS, MN was obtained using 4 matched peaks within a m/z tolerance of 0.02 Da and minimal cosine value of 0.72; the maximum neighbor number (topK) was set to 10 and the maximum number of connected component size at 1000. Filtering was operated for peaks outside the ± 17 Th window and by keeping each peak in the top 6 in the ± 50 Th window. For t-SNE construction, at least 1 cosine scores above 0.75 were searched by operating 1000 iterations with perplexity 6, learning rate 200, and early exaggeration 12. The Barnes-hut approximation with an angle of 0.5 was used. Parameters for MN construction were chosen by preliminary testing of different combinations (not shown). The best ones for our study resulted from a compromise for the best clusterization of both compounds which fragmented extensively and modestly. Molecular networking was also performed on the GNPS website in order to confirm results (not shown), while public access to the GNPS projects can be found at: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=9ec810a23cd043c3806987b2c524789c for positive mode and at: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3396379174ab4cfba323fdfa324b6e18 for negative mode.
The mzXML centroided files were uploaded on the Sirius platform (version 5.8.3) [73] for molecular formula assignment, which was chosen as the most probable calculated according to the combination of both the Molecular Formula Identification tool and the Fingerprint prediction and Structure database search CSI:FingerID tools [74, 75]. For the first, the Q-TOF instrument was chosen, using a mass accuracy of 5 ppm and atom constraints 0 < H < ∞, 0 < C < ∞, 0 < N < ∞, 0 < O < ∞, 0 < P < 2, 0 < S < 2. Filtering was used for the MS1 isotope pattern and the MS/MS scorer. Ten candidates for ions were stored, with a minimum of 1 candidate for ion. Possible ionizations were [M + H]+, [M + Na]+, and [M + K]+. For CSI:FingerID, fallback adducts used for fingerprint prediction were [M + H]+, [M + Na]+, [M + K]+, [M + NH4]+, and [M-H2O + H]+. All proposed databases were used for structure database search.
Compound annotation was performed on classic CID fragmentation spectra based on database search on MetGem and GNPS by searching 3 fragments and setting a m/z tolerance of 0.02 Da with a minimal cosine score of 0.65 and m/z parent filter tolerance at 17Th. Analogs were searched in a maximum span of 100 Da difference from precursor ions. Compounds which were not successfully annotated through databases were further investigated and annotated through a study of the obtained molecular families, the t-SNE and manual annotation on the fragmentation spectra and on the information obtained on the Sirius platform. Regarding MN constructed for phosphatidylcholines using DDA-EAD-MS spectra, parameters for MN construction remained unvaried.
Results and discussion
Gammarus fossarum metabolome exploration by classic DDA-CID-MS fragmentation and molecular networking
G. fossarum extracts (C1 and D1 reproductive stages) were analyzed through DDA-CID-MS in both positive and negative ionization mode. Representative base peak intensity (BPI) and total ion current intensity (TIC) chromatograms obtained from these analyses are shown in Figures S1 and S2. An observation of these BPIs points out on a higher number of metabolites readily detected in positive ion mode with respect to the negative one and also, on a relatively higher intensity of non-polar compounds with respect to the more polar ones. Data analysis restituted 823 features (defined as detected ions associated to an experimental m/z and tR values) in positive ion mode and 98 in negative ion mode, present in at least two analytical replicates for each reproductive stage (details on data handling and processing are given in the material and method section). The final feature lists were then submitted to molecular networking construction on MetGem (version 1.3.6). The MN obtained in negative ion mode (Figure S3) resulted limited and compound annotation mainly confirmed results obtained in positive ion mode. For this reason, the next discussion will be focused on the positive ionization mode. Figure 1 shows the MN obtained in positive ion mode with some representative compounds annotated within the obtained molecular families. MNs allowed together with molecular formula attribution and database search, to obtain the annotation of 55% of the total features at level 2, 19% at level 3, and 26% at level 4 for the positive ion mode [76]. On note, while in-source fragments were discarded for MN construction, adducts were kept improving the annotation of certain molecular classes, such as the TGs (detected solely as ammonia adducts). The 823 features corresponded to a total of 742 unique compounds. The complete list of features detected in the G. fossarum extracts together with their annotation, relative intensity, and annotation confidence level can be accessed as supplementary file (Feature_list_Gfossarum). Several molecular families at a high number of connected nodes corresponded to lipids. Among those, the most represented lipids were monoglycerides (MGs), phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), plasmalogens (PC-Os and PE-Os), phosphatidylinositols (PI), and carnitines (CARs). Other annotated lipids were fatty acids (FAs), diacylglycerols (DGs), triacylglycerols (TGs), phosphatidylserines (PSs), lysophosphatidylcholines (LPCs), and lysophosphatidylethanolamines (LPEs). PEs and PSs gathered in the same molecular family corresponded to protonated forms of lipids having the same side chains. In fact, the neutral loss of the phosphoethanolamine (141.06 Da) and the phosphoserine (185.07 Da) polar heads, respectively for PEs and PSs, led to fragmentation spectra which presented mainly the same fragments.
Full molecular networking obtained in positive ionization mode for G. fossarum extracts analyzed through DDA-CID-MS conditions with representative examples of annotated metabolites for each molecular family. Node color represents the semi-quantitative analysis performed by comparing the relative intensity of each feature in the C1 (light blue) and D1 (red) reproductive stages
In addition, three molecular families gathered features which matched with taurolipids, i.e., peculiar lipid classes characterized by a sulfur atom in the form of a sulfone functional group. In particular, the first family (e in Fig. 1) was constituted of N-acyl taurines (TAUs), bioactive lipids which have been found to contribute to glucose homeostasis and lipid metabolism and their existence has already been proved in different marine invertebrates and crustaceans [77, 78]. Similarly, molecular family l gathered molecules that resulted to be analogs of taurolithocholic acids, particular classes of bile acids in humans. The third molecular family of taurolipids (o in Fig. 1) was constituted of only two nodes and was represented by two features at m/z 590.4806 [C33H67NO5S + H]+ and m/z 620.4909 [C34H69NO6S + H]+. These lipids have been already encountered in the hepatopancreas and the caeca of G. fossarum females in a previous study, but their fragmentation spectra had restituted an unclear annotation [65]. Fragmentation mass spectra obtained in positive ion mode showed characteristic neutral losses of the acyl chain and of the TAU chain and also a fragment at m/z 124.00 attributed to [C2H5NO3S + H]+. Their experimental fragmentation spectra agreed with reference data for sulfonolipids, a unique class of sphingolipids featuring a sulfonate group and containing an N-acyl taurine as a side chain.
Despite a high percentage of the detected compounds in the analyzed extracts corresponding to lipids, G. fossarum extracts showed many primary and secondary metabolites. In this case, the principal molecular family included different compounds involved in the energy and/or nucleic acid metabolism, as nucleosides and nucleotides; or di- and oligopeptides containing arginine. In general, other metabolite families contained simple amino acids, as glutamic acid, tyrosine, phenylalanine, or sugars, glucose, maltotriose, and lactotriose.
Other remarkable metabolites were found as smaller families or as single nodes. Among those, feature 126 (in table Feature_list_Gfossarum) at m/z 292.0554 was attributed to Se-methyl selenoneine and feature 14 to a potentially new Se-methyl selenoneine analog still not present in current literature and likely containing an amide functional group. For these compounds, structure assignment was performed according to a comparison of experimental and predicted isotopic pattern distribution, molecular formula assignment, and manual fragmentation spectrum rationalization (Figure S4). Compounds containing selenium have shown high interest in ecotoxicology because of their supposed fundamental role in heavy metal and free radical scavenging [79, 80]. Other metabolites with presumed protective function were found in G. fossarum extracts as for example compound 44 at m/z 347.1449 and molecular formula [C14H22N2O8 + H]+ which matched to Porphyra-334 (IUPAC name 2-[[3-(carboxymethylimino)-5-hydroxy-5-(hydroxymethyl)-2-methoxycyclohexen-1-yl]amino]-3-hydroxybutanoic acid), an ultraviolet filter produced by cyanobacteria and which has been found in some crustaceans [81].
DDA-CID-MS conditions applied on G. fossarum extracts enabled a first annotation thanks to database match or to manual fragmentation spectrum inspection for most of the detected features. In the attempt to find a possible structure attribution to additional compounds which remained unknown, the t-SNE algorithm on MetGem was used to find a link among groups of families with structural similarities. Figure S5 shows the relationships found between the classic MN and the obtained t-SNE in positive ionization mode. This last showed three zones of interest which regrouped molecular families not connected in the classic MN and which had not been yet annotated. In particular, the TAUs molecular family associated to compounds which likely contained one or two additional oxygen atoms according to molecular formula assignment. Those related compounds were tentatively annotated as compounds in which the polar taurine head was subjected to acetylation and di-acetylation or to oxidized species. Confirmation of the proposed TAU analogs should be obtained through additional experiments, including compound isolation and fine structure characterization. The green and orange zones on the t-SNE regrouped PCs and LPS, respectively as H+ and Na+ adducts, and included some molecular families at a low number of nodes which were not linked together by the classic MN.
Figure 1 proves the presence of certain molecular families which nodes have a predominant red color, which would indicate a relatively higher intensity of the feature at the D1 stages. Among these, the TAUs, the taurolitocholic acid analogs, and in less extent compounds involved in energy metabolism seem to be specifically expressed at this stage, implying a particular role in molting and/or female gonadic development.
On another hand, this study led to the discovery of compounds which are probably introduced in G. fossarum metabolism thanks to its diet and the interaction between diet and the symbiont microbiome of these organisms [82]. For example, sulfonolipids have been proven to be of bacterial origin [83] and have been recently discovered in mice and to be associated to high-fat diets [84] and to trigger multicellular development in living relatives to animals [85]. Protective metabolites such as Porphyra-334 or carotenoids such as astaxanthin and its analogs are normally produced by plants and may be introduced in G. fossarum diet by algae ingestion. In addition, a high number of metabolites and lipids containing sulfur has been found, supporting their involvement in many essential biological roles in amphipods and, on the other hand, their responsibility in the flavor of many aquatic organisms [86, 87].
General comparison of the CID and EAD fragmentation spectra for the annotated metabolites in G. fossarum female extracts
EAD fragmentation pathways have been thoroughly studied for specific classes of lipids such as PC, DG, and TG and for specific classes of metabolites, such as nucleosides, pharmaceuticals, or agrochemicals [45,46,47,48, 50,51,52,53]. Nevertheless, many other classes of compounds have been omitted from a detailed explanation of their behavior during fragmentations under EAD conditions. In general, no universal rules have been extrapolated for EAD fragmentation which could explain the fragmentation pathway of compounds of different natures. As described in the previous paragraph, G. fossarum extracts analyzed in this study presented many different classes of lipids and metabolites with various structures, which are well suited to perform a systematic comparison of the two fragmentation modes and the information delivered for compound annotation. In EAD, ion activation through the impact with multiple electrons in the collision cell results in a higher scan time required for the acquisition of a single spectrum with respect to a CID fragmentation event. In this study, DDA on TOP10 peaks was kept in the two fragmentation modes by adapting the EAD reaction time at 30 ms. These parameters assured similar total scan times for DDA-CID-MS and DDA-EAD-MS conditions, resulting respectively in 1.238 s and 1.255 s, which led to a total of 1350 and 1150 fragmentation events respectively obtained as a mean value on three chromatographic runs of 34 min for one case sample. As claimed before, a final list of 823 unique features which fragmentations had been acquired in both conditions (CID and EAD) was obtained and used for classic analysis (see paragraph 3.1) and for further EAD studies. Once annotation based on classic methods was performed, a first general comparison between CID and EAD fragmentations was performed according to a simple rule: following the number of fragment ions formed under the two conditions for each compound. While this could appear as a rough parameter, the number of formed fragments of a compound eventually influences the quality of the annotation and the possibility of assigning substructures or discriminating structural isomers. It is worth saying that before counting the number of formed fragments, to discriminate relevant fragment peaks from noise and obtain a robust comparison among CID and EAD fragmentations, the level of noise was first studied. In the absence of a preliminary differential mobility spectrometry (DMS) ion separation, noise in CID fragmentation spectra obtained through a Q-TOF depends mostly on the relatively large isolation window of the selecting quadrupole (in this study unitary mass, i.e., ± 0.35 Da), which can produce low-intensity satellite peaks around the selected precursor mass which undergo fragmentation at the same time, especially in the case of complex samples presenting co-eluting isobars [48]. Moreover, the intensity of fragments related to satellite peaks could be increased because of the Zeno Trap pulsing signal enhancement. Regarding EAD fragmentation, in addition to the aforementioned reasons, additional noise may arise from the residual gas molecules which are ionized by the electrons [47]. In order to reduce background peaks, the noise level was first checked manually using a set of 20 fragmentation spectra having base peak intensity (BPI) in the range of 1·102 to 1.6·106 counts, and then, background intensity collected setting the minimal intensity at a value accounting for confident fragment peak assignment. In our study, the noise level in fragmentation spectra resulted in a comparison between CID and EAD, probably thanks to the noise background reduction step performed before the acquisition of the DDA-EAD-MS injections. Importantly, the noise level in the two fragmentation modes resulted similarly influenced by the overall base peak intensity, which depended primarily on the number of isolated ions and from the amount of fragmented precursor ions. Collected values highlighted a linear correlation between noise and MS/MS BPI values on a logarithmic scale (Figure S6) which was hence used to set a minimal intensity threshold for fragment peak picking in the MS/MS spectra for comparison in CID and EAD conditions. This assured also that small differences in the BPI of the fragmentation spectra for the same compound under CID and EAD would not lead to wrong comparisons of the formed fragments.
A selection of 331 out of the 823 total features was chosen for comparison. This list included compounds at a high annotation confidence level (level 2 or level 3 supported by confident fragmentation spectra interpretation). For each compound, the number of fragments formed in CID and EAD conditions was then obtained according to the threshold relative to the fragmentation spectra BPI. Figure 2a shows the mirrored number of fragments formed in CID and EAD conditions for the 331 compounds disposed of according to a growing m/z order, while the difference between the two values is shown in Fig. 2b. Despite the noise subtraction, a relatively high number of fragments (on average 50–100) were obtained for the entire set of studied compounds, in both CID and EAD conditions. Nevertheless, it must be considered that the count of peaks included, together with high-intensity fragments, additional fragments not generally present in literature conditions, which intensity has been increased by the activation of the ZenoTrap pulsing. Considering that the comparison was performed for the same compound in different conditions, the peak redundancy should not influence the overall conclusions.
Comparison of the formed fragments for the features of this study relative respectively to a the number of fragments formed for each compound (on a selection of 331 out of the final 823, formed in EAD (magenta) and CID (black) conditions); b their formal subtraction; c the number of fragments formed for the 823 compounds of this study obtained in EAD (magenta) and CID (black) conditions in m/z windows of 10 Da; and d the mean intensity of these fragments
Figure 2b shows that except in sporadic cases, the number of fragments obtained in EAD becomes consistently higher than that in CID conditions only at higher m/z ratios (from m/z 500 to 1200). Moreover, most of the compounds at high m/z corresponded to lipids, for which fragmentation through EAD at 10 eV has already proven to be extensive. Nonetheless, other compounds at high m/z for example oligopeptides and polysaccharides produced a higher number of fragments in EAD with respect to the applied CID conditions. Contrarily, only 36% of the selected features at lower m/z (up to 500) presented additional fragmentation in EAD mode, as residual CID fragments stood out in those spectra.
One reason could be that the larger the ion size, the greater the available ion surface for colliding with electrons during EAD. A more compelling explanation relies on the mechanisms of the two dissociations. In CID, commonly characterized as a thermal dissociation process, an initial activation step converts the translational energy of the accelerated ion into internal energy. This internal energy is then randomly distributed among all the vibrational modes of the ion, leading to bond cleavage, as per the quasi-equilibrium theory (QET) [88]. Given that the number of vibrational modes increases with molecular mass, maintaining the same collision energy can result in decreased ion yields, as some bonds may not acquire sufficient energy for cleavage [30]. In contrast, EAD involves a distinct activation process mediated by high-energy electrons. These electrons cause the excitation of the electronic states of the molecular ion, resulting in a different amount of acquired internal energy [44].
It can be speculated that the electronic excitation induced by EAD was effective in causing bond cleavage for most compounds at higher m/z compared to the increase in internal energy caused by the applied CID conditions in this study.
In a second step, all fragment ions formed for all the 832 compounds were gathered without assignment to the related precursor ion and ordered according to their growing m/z value. The number of produced fragments in windows of 10 m/z along the full acquisition range (50–1200 Da) and their intensity was then studied for the two fragmentation modes. Figure 2c shows that CID fragmentation led, for the compounds of this study, to a higher number of fragments at low m/z value (up to m/z 200–250), while EAD produced, in absolute, fragments which well distributed in all the m/z range, highlighting the possibility to obtain additional peaks accounting for fine substructure assignment. As the ZenoTrap pulsing was used for both fragmentation modes, this difference should be attributed to the specific mechanisms of CID and EAD fragmentations for the type of compounds analyzed in our samples. Despite a generally higher number of fragments formed in DDA-EAD-MS mode, the mean intensity resulted much lower in the case of EAD (Fig. 2d), with most of the peaks falling near the limit of the background noise and reducing the overall confidence for peak interpretation and structure annotation. On note, fragmentation spectra quality could be improved by increasing the reaction time or the accumulation time, at the expense of the number of acquired fragmentation events.
In conclusion, it is noteworthy that some of the fragments counted in the EAD spectra might originate from CID-type fragmentations due to the minimal collision energy (CE) set in the collision cell (10 eV). The differentiation between common and additional fragments was not considered in the construction of the previous graphs. This consideration would have demanded a meticulous examination and comparison of the CID and EAD spectra for each investigated compound. Therefore, the subsequent paragraph is dedicated to a more in-depth discussion of the additional fragments formed under EAD conditions for representative compounds from the principal molecular classes.
CID and EAD fragmentation pathways for the principal classes of compounds annotated in G. fossarum female extracts
To obtain more detailed information, EAD fragmentations were studied for single compounds in an attempt to confirm or improve the annotations performed through CID spectra. The most intense ion for the main molecular classes obtained in the MN was selected as case examples to discuss the fragmentation mechanisms.
G. fossarum extracts showed a high richness of PCs, with 67 nodes constituting the obtained molecular family. Among these, the most intense ion at m/z 760.5826 corresponding to [C42H82NO8P + H]+ fragmented under CID by producing the two fragments at very low intensity at m/z 522.3539 and at m/z 496.3398, which corresponded to a PC 18:1_16:0. The EAD spectrum for this compound (Figure S7-a) presented the fragment ions at m/z 224.1056 and m/z 226.0838 which allowed excluding the presence of a sphingomyelin and the fragments at m/z 479.31 and 505.3295 which confirmed fatty acid positions and additional side chain radical fragmentations which enabled locating the double bond at the n-9 position. Similarly, the EAD fragmentation spectra of selected PEs (PE 18:1_20:5 at m/z 764.5247), LPCs (LPC 18:1 at m/z 522.3549), and LPEs (LPE 18:1 at m/z 480.3080) confirmed and improved structure assignment, highlighting the presence of n-6, n-8, and n-4-based lipids and enabling double bond position assignment even in the case of polyunsaturated side chains, as shown in Figure S7-b, S7-c, and S7-d. For LPEs, fatty acid radical cleavages seemed to take place preferentially after the loss of a water molecule (fragment ion at m/z 462.2969 in Figure S7-b), as no fragments could be associated to a direct fragmentation of the protonated precursor ions. In general, for all the compounds belonging to these molecular classes, the EAD fragmentation spectra could easily be used to retrieve double bond position on the fatty acid chains, when BPI intensity resulted higher than 5·103. Regarding double bond stereochemistry, attributions could not be confidently performed, even if peak intensity variation for radical and hydrogen losses concomitant to double bonds suggested a prevalence of cis configurations. The comparison with references is advised to obtain a more accurate insight into double bond configuration. For FAs, MGs, DGs, and TGs, EAD spectra presented intense fragment peaks in common with the CID spectra (Figure S8) and additional radical cleavages of the acid fatty chains. These last enabled, in the case of the studied MG, to speculate on the presence of branched side chains and locate them. Even if the most intense DG and TG were selected, information on double bond localization or conformation could not be accessed because of the poor spectra quality and the presence of coeluted species.
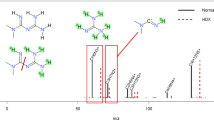
For CARs, the compound at m/z 398.3265 corresponding to [C23H43NO4 + H]+ was the O-acylcarnitine 16:1. The EAD fragmentation spectra showed in Fig. 3a showed a principal fragment arising from the loss of a radical acyl group from the protonated precursor ion. All the other additional fragments were likely formed from the concomitant radical cation at m/z 397.3186 and a [M − H]+ ion at m/z 396.3105, present in the MS/MS spectrum together with the precursor ion. Moreover, additional fragments seemed to be generated by the formation of oligomers produced by the reaction among the formed radicals during reaction time. As an example, the fragment ion at m/z 338.9666 may be obtained as the combination of the acyl radical (44.99 Da) and other radical species formed during the EAD fragmentation. The EAD fragmentation spectrum did not show any pattern associable to successive cleavages of the side chain induced by radical losses and therefore it was not possible to obtain additional information on double bond localization or conformation. In the case of TAUs, the CID and EAD fragmentation spectra for the feature at m/z 390.2670 corresponding to the molecular formula [C20H39NO4S + H]+ were selected for discussion. The annotation on CID spectra was based on the presence of the diagnostic peak at m/z 126.0219 [C2H7NO3S + H]+ corresponding to the protonated taurine head group and, in addition, on the release of the protonated alkyl chain with successive neutral loss of water. The corresponding EAD fragmentation was characterized by fragments corresponding to the cleavage of the sulfonic head group with the release of OH▪ or HSO3▪. More importantly, two additional fragments at m/z 182.0482 and m/z 167.0247, which were not present in the CID spectra, likely corresponded to the cleavage of the alkyl chain in the proximity of the double bond. Therefore, while the CID fragmentation allowed annotation as TAU 18:1, EAD spectra enabled a tentative localization of the unsaturation, as shown in Fig. 3b.
Mirrored EAD (magenta) and CID (black) fragmentation spectra for the most intense a CAR, b TAU, c SL, and d carotenoid (astaxanthine) detected in G. fossarum extracts. For each compound, a proposition of structure performed on both CID and EAD spectra and fragment rationalization has been added. Panel a shows a zoom on concomitant peaks (m/z 397.3172 and m/z 396.3084) formed together with the precursor ion at m/z 398.3261. Spectra have been zoomed to reach equal intensity for both CID and EAD conditions
For SL, the ion at m/z 620.4909 corresponding to [C34H69NO6S + H]+ fragmented in CID conditions by forming the fragment at m/z 124.006 (taurine head group) and fragments corresponding to the releases of an acyl-sulfone (m/z 286.2741) and of an acyl-amide (m/z 352.2516), followed by water loss. Those main fragmentations were present also in the EAD spectrum, together with some additional fragments arising from the loss of the radical sulfone (m/z 539.5236), from the radical sulfone plus a radical hydroxy group (m/z 520.5032) and from this last species of a radical unsaturated alkyl chain (possible structure shown in Fig. 3c). The selected SL was tentatively annotated as sulfobacin A, which according to literature data contains two branched fatty acids. Fragment peaks observed in both CID and EAD spectra did not provide any information in this regard, preventing confirmation of the performed annotation. The proposed annotation for features 695 (Feature_list_Gfossarum) at m/z 597.3912 and molecular formula [C40H52O4 + H]+ was astaxanthin. A confidence level 3 annotation was obtained for this compound based on manual CID and EAD fragmentation mass spectra investigation. In fact, Fig. 3 shows that additional fragments in EAD were likely formed by radical cleavage of the unsaturated linear chain, strengthening the annotation performed on the classic CID method. Nonetheless, many other EAD fragments could not be rationalized according to the mere structure of astaxanthin, leading to the possible presence of a different structural isomer.
Most of the metabolites formed EAD fragments which resulted in specular radical species of the CID ones, as shown in Figure S9 for porphyra-334, Se-methyl selenoneine, and riboflavin. Additional EAD fragments, when present, could be used to confirm CID-based annotations and obtain an insight into the peculiar functional group connection for metabolites. For example, the loss of a radical methyl group from the Se-methyl selenoneine parent ion, which was obtained exclusively in EAD conditions, helped confirm its bond with selenium (Figure S9-b). Nevertheless, in some cases, EAD additional peaks could not be easily attributed on the basis of simple radical bond cleavages of the protonated precursor ion, as shown in Figure S9-c. The formation of these fragments was observed for metabolites containing carbonyl, carboxyl, or sulfonyl functional groups and it could be due to the combination of radical oxygen species released from the protonated precursor ion with other ionic/radical species, during reaction time or during their path within the instrument.
Importantly, EAD fragmentation for both metabolites and lipids was performed using a fixed kinetic energy of 10 eV, as the DDA-MS mode in EAD conditions does not allow at present to dynamically adapt the applied energy during the acquisition. Higher values of kinetic energy will be investigated in the future to obtain more informative compound fragmentation spectra. In the optic of a global compound annotation based on DDA-MS (EAD), we attempted to confirm and improve the CID annotations by comparing them with the EAD experimental spectra for each compound. EAD fragmentation spectra generally led to more confident structure annotations, in particular for lipids and in the case of metabolites for which a hypothesis of structure had been obtained through preliminary CID, while the full annotation of unknown species based on EAD remained very complex.
Towards the use of molecular networking based on DDA-EAD-MS conditions for the faster annotation of lipid species
Considering that lipid species fragment under EAD conditions by delivering more informative spectra, we attempted the construction of molecular networking based on DDA-EAD-MS analysis. A comparison of the molecular family obtained in CID and EAD conditions for the PCs detected in G. fossarum samples was performed. Text files containing fragmentation peaks and relative intensity were manually created by putting particular attention to the correspondence of m/z of the precursor ions, retention time, and assigned feature number for each feature, in the two conditions. The two molecular networking were then constructed by using the same parameters and compared as shown in Fig. 4. In order to highlight correlations or differences, annotation was performed manually according to their experimental CID and EAD spectra, and the corresponding features highlighted in different colors, by following the different positions in a particular zone of the EAD molecular networking. A visual inspection of Fig. 4 highlights that compounds are differently connected among them in the CID and the EAD-obtained molecular networking. In fact, in CID conditions, the only fragment present in all the fragmentation spectra of PCs was the ion at m/z 184.07, and node connections in CID MN were therefore created according to the intensity variation of this fragment. On the other hand, taking that the EAD fragmentation spectra present additional fragments arising from the fatty acid cleavages, the corresponding MN resulted in more precise node connections to be used for PC description. In particular, while the two additional fragments at m/z 224.104 and m/z 226.084 formed in EAD conditions and discriminant for PCs resulted not discriminant because they were present in all fragmentation spectra, PC clustering in EAD conditions seemed to be principally influenced by fragments formed by cleavage of the glycerol portion. Hence, PCs with C18:1 fatty acids clustered more tightly (pink nodes in Fig. 4) thanks to the presence of diagnostic fragments at m/z 465.322 and m/z 505.306. Manual assignment for each PC highlighted that PCs in the pink zone also presented a majority of double bonds in positions n-3 and n-9. However, it is not possible to determine if the prevalence of n-3 and n-9 bonds in the pink zone arose from the natural distribution of lipids in G. fossarum samples or because of the presence of common characteristic fragments and neutral losses.
Molecular networking obtained for the molecular family of PCs found in G. fossarum samples in a CID and b EAD conditions. Compounds corresponding to a particular zone in the two representations have been evidenced with different colors. Each node shows the measured mass and the attribution performed manually on the basis of the a CID or b EAD spectra. Semi-quantitative analysis between C1 and D1 features as well as node size has been neglected
For example, let’s compare two compounds grouped within the pink zone (m/z 760.584 annotated as PC 18:1_16:0 (n-9), 0.6 ppm, and m/z 746.5692 annotated as PC 18:1_15:0 (n-9), 1.6 ppm). In the event of a radical fragmentation adjacent to an n-9 double bond on PC 18:1_16:0, a relatively intense fragment at m/z 661.4677 (C35H68NO8P•+) is expected, possibly due to electron stabilization through conjugation. Conversely, a similar fragmentation event on PC 18:1_15:0 results in a relatively intense fragment at m/z 647.4521 (C34H66NO8P•+). In both cases, a common feature is the radical loss of 99.117 Da (C7H15•).
Additionally, some fragments arising from cleavages in the side chains of the two PCs at different points can share the same m/z values, even though they may not necessarily correspond to identical species (see Figure S10). These fragments can contribute to an increase in the final cosine score for a feature pair. The presence of PCs with different compositions in a particular zone may thus be explained by the existence of shared m/z values, even if they do not precisely reflect structural similarities.
Similar results were obtained for the yellow zone and the green zone in the EAD molecular networking, gathering respectively PCs showing fragments thanks to the presence of diagnostic fragment at m/z 525.322, which led to the presence of C20 fatty acids with five unsaturation and a majority of double bond in position n-3, n-6, n-9, n-12 and n-15 and PCs with a predominance of C22 fatty acids and six unsaturations with a preferential double bond localization in positions n-3, n-6, n-9, n-12, n-15, and n-17 (fragment at m/z 551.337). Those results agree with previous shotgun lipidomics studies performed on G. fossarum organisms [65]. In any case, it was not possible to extrapolate, as an absolute, fragments that are in common among all PCs for a zone, and EAD clustering seemed to be a result of a complex balance among common m/z values and neutral losses. However, manual annotation of nodes in the three zones of the EAD molecular family showed that somehow EAD MN succeeded in reaching a deeper level of clustering compared to CID MN. Additional studies are highly needed for a better comprehension of clustering under EAD conditions. Moreover, the EAD molecular networking showed novel single nodes, i.e., compounds which were disconnected from the rest of the molecular family. This disconnection with respect to the CID molecular family could be explained by the very low intensity of fragments indicative of double bond localization. It is worth noting that increasing spectra quality, for example, by increasing the reaction time or the accumulation time, should lead to improved connection on EAD-constructed MN.
Many attempts performed on the comparison of MNs in EAD and CID conditions for molecular families containing smaller molecules as metabolites or constructed for the full set of 823 selected features have led to inconclusive results. Nevertheless, MN constructed based on EAD fragmentation spectra showed to be a very promising tool for the aided annotation of lipids with respect to the classic MN constructed under CID conditions, even if some efforts need to be put into manual spectra assignment.
Conclusion
In the present study, we provided a comprehensive characterization of metabolites and lipids detected in entire organisms of G. fossarum females at two reproductive stages by using DDA-CID-MS and DDA-EAD-MS fragmentation conditions. We started from a classic approach which enabled us to compile a list of 823 annotated features through molecular networking and t-SNE construction, database search, and manual interpretation based on CID spectra. G. fossarum analyzed extracts were confirmed to be very rich in lipids, containing many of the widespread classes such as PCs, PEs, CARs, and DGs, to name a few. These results agreed with previous studies carried out on G. fossarum lipidomics analysis [65, 89]. In addition, less expected lipids containing sulfur as TAUs, SL (sulfonolipids), and taurolithocholic acids were annotated, and two potential new classes of oxidated TAUs discovered. Many primary and exogenous metabolites were also detected in G. fossarum samples, for example, nucleosides, nucleotides, sugars, amino acids, proteins, chitin residues, and molecules which probably have defensive effects as heavy metal chelators, UV filters, and scavenging activity. In future studies, extraction of G. fossarum whole organisms will be carried out using alternative solvents in order to possibly broaden the range of detected and studied compounds up to more complex structures.
In order to give more confidence to the annotations performed in the case of a non-model organism for which metabolites and lipids are still not fully elucidated, we conducted a critical evaluation of the use of DDA-EAD-MS condition, as an alternative fragmentation mode. As a first example in the literature on the use of DDA-EAD-MS for comprehensive compound annotation, we first performed a critical study of the two acquisition modes by comparing the noise variation in function of the fragmentation spectra BPI in the two dissociation modes, the number of fragments generated in CID and EAD for each compound, and their variation in terms of number and intensity over the acquired m/z range. Moreover, we performed a specific comparison and rationalization of the CID and EAD fragmentation pathways for case compounds belonging to the principal molecular classes detected in G. fossarum extracts. EAD resulted in a general increase of the formed fragments at increasing precursor ion size and in the formation of a higher number of fragments well distributed over the full m/z range with respect to CID, which however led to fragments at an overall higher intensity. Deep investigation of the CID and EAD spectra confirmed EAD a better choice for the annotation of “classic” lipids as PCs and PEs, as it gave access to quasi-complete structural characterization up to double bond position information. For most of the other investigated species, EAD spectra enabled us to confirm the annotations performed on the basis of CID and speculate on additional structural features, such as the presence of branched chains. On note, EAD failed to be more informative than CID for some species as carnitines or particular metabolites, and additional EAD fragments could not always be assigned to real radical cleavages but to the combination of multiple radical species within the instrument. Finally, we built molecular networking for the phosphatidylcholines detected in the study using EAD fragmentation spectra and performed a comparison highlighting complex connections between nodes for phospholipids since more fragments are produced in positive ion mode under EAD. On the other hand, the attempts to compare MNs under CID and EAD, whether focused on molecular families with smaller metabolites or encompassing the entire set of 823 selected features, have yielded inconclusive results.
On another side, this study may suffer from some limitations, for example, the impossibility of performing DDA-EAD-MS using dynamic KE or KE spread for optimized compound fragmentation which introduced a bias for comparison with DDA-CID-MS performed with CE spread conditions. Moreover, despite our efforts, the negative ionization mode could not be exploited, because of the unsatisfactory number of detected features in G. fossarum samples. The next investigations performed in our group will be dedicated to the exploration of this aspect, which is very poorly discussed in the published literature.
In conclusion, DDA-EAD-MS has been revealed as a powerful complementary tool to classic DDA-CID-MS to gain high-throughput information on features in complex biological samples. Importantly, the bottleneck of the use of EAD still lays on a one-to-one manual spectra interpretation, together with the lack of reference data in the public databases and of complete studies delivering EAD fragmentation rules. We think that, in the future, further efforts should be made towards shared EAD fragmentation libraries and rules and towards the optimization of programs and tools for improved EAD-based compound annotation.
Data availability
All the data are described within the manuscript. The raw data and metadata analyzed during the current study are available from the corresponding author on request.
References
Vailati-Riboni M, Palombo V, Loor JJ. What are omics sciences? In: Periparturient diseases of dairy cows. Cham: Springer International Publishing; 2017. p. 1–7.
Wang R, Li B, Lam SM, Shui G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression. J Genet Genomics. 2020;47:69–83. https://doi.org/10.1016/j.jgg.2019.11.009.
Gallart‐Ayala H, Teav T, Ivanisevic J. Metabolomics meets lipidomics: assessing the small molecule component of metabolism. BioEssays. 2020;42. https://doi.org/10.1002/bies.202000052.
Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol. 2018;36:316–20. https://doi.org/10.1038/nbt.4101.
Heiles S. Advanced tandem mass spectrometry in metabolomics and lipidomics—methods and applications. Anal Bioanal Chem. 2021;413:5927–48. https://doi.org/10.1007/s00216-021-03425-1.
Rakusanova S, Fiehn O, Cajka T. Toward building mass spectrometry-based metabolomics and lipidomics atlases for biological and clinical research. TrAC Trends Anal Chem. 2023;158:116825. https://doi.org/10.1016/j.trac.2022.116825.
Ribbenstedt A, Ziarrusta H, Benskin JP. Development, characterization and comparisons of targeted and non-targeted metabolomics methods. PLoS One. 2018;13:e0207082. https://doi.org/10.1371/journal.pone.0207082.
Gertsman I, Barshop BA. Promises and pitfalls of untargeted metabolomics. J Inherit Metab Dis. 2018;41:355–66. https://doi.org/10.1007/s10545-017-0130-7.
Deschamps E, Calabrese V, Schmitz I, Hubert-Roux M, Castagnos D, Afonso C. Advances in ultra-high-resolution mass spectrometry for pharmaceutical analysis. Molecules. 2023;28:2061. https://doi.org/10.3390/molecules28052061.
Chaleckis R, Meister I, Zhang P, Wheelock CE. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr Opin Biotechnol. 2019;55:44–50. https://doi.org/10.1016/j.copbio.2018.07.010.
Guo J, Huan T. Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography–mass spectrometry based untargeted metabolomics. Anal Chem. 2020;92:8072–80. https://doi.org/10.1021/acs.analchem.9b05135.
Wang R, Yin Y, Zhu Z-J. Advancing untargeted metabolomics using data-independent acquisition mass spectrometry technology. Anal Bioanal Chem. 2019;411:4349–57. https://doi.org/10.1007/s00216-019-01709-1.
Barbier Saint Hilaire P, Rousseau K, Seyer A, Dechaumet S, Damont A, Junot C, Fenaille F. Comparative evaluation of data dependent and data independent acquisition workflows implemented on an Orbitrap Fusion for untargeted metabolomics. Metabolites. 2020;10:158. https://doi.org/10.3390/metabo10040158.
Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12:523–6. https://doi.org/10.1038/nmeth.3393.
Yin Y, Wang R, Cai Y, Wang Z, Zhu Z-J. DecoMetDIA: deconvolution of multiplexed MS/MS spectra for metabolite identification in SWATH-MS-based untargeted metabolomics. Anal Chem. 2019;91:11897–904. https://doi.org/10.1021/acs.analchem.9b02655.
Stancliffe E, Schwaiger-Haber M, Sindelar M, Patti GJ. DecoID improves identification rates in metabolomics through database-assisted MS/MS deconvolution. Nat Methods. 2021;18:779–87. https://doi.org/10.1038/s41592-021-01195-3.
Tada I, Chaleckis R, Tsugawa H, Meister I, Zhang P, Lazarinis N, Dahlén B, Wheelock CE, Arita M. Correlation-based deconvolution (CorrDec) to generate high-quality MS2 spectra from data-independent acquisition in multisample studies. Anal Chem. 2020;92:11310–7. https://doi.org/10.1021/acs.analchem.0c01980.
Alseekh S, Aharoni A, Brotman Y, Contrepois K, D’Auria J, Ewald J, Ewald CJ, Fraser PD, Giavalisco P, Hall RD, Heinemann M, Link H, Luo J, Neumann S, Nielsen J, Perez de Souza L, Saito K, Sauer U, Schroeder FC, Schuster S, Siuzdak G, Skirycz A, Sumner LW, Snyder MP, Tang H, Tohge T, Wang Y, Wen W, Wu S, Xu G, Zamboni N, Fernie AR. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat Methods. 2021;18:747–56. https://doi.org/10.1038/s41592-021-01197-1.
Hoskisson PA, Seipke RF. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. mBio. 2020;11. https://doi.org/10.1128/mBio.02642-20.
Little JL, Cleven CD, Brown SD. Identification of “known unknowns” utilizing accurate mass data and chemical abstracts service databases. J Am Soc Mass Spectrom. 2011;22:348–59. https://doi.org/10.1007/s13361-010-0034-3.
Little JL, Williams AJ, Pshenichnov A, Tkachenko V. Identification of “known unknowns” utilizing accurate mass data and ChemSpider. J Am Soc Mass Spectrom. 2012;23:179–85. https://doi.org/10.1007/s13361-011-0265-y.
Perez De Souza L, Alseekh S, Brotman Y, Fernie AR. Network-based strategies in metabolomics data analysis and interpretation: from molecular networking to biological interpretation. Expert Rev Proteomics. 2020;17:243–55. https://doi.org/10.1080/14789450.2020.1766975.
Phelan VV. Feature-based molecular networking for metabolite annotation. 2020;227–243.
Nothias L-F, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, Protsyuk I, Ernst M, Tsugawa H, Fleischauer M, Aicheler F, Aksenov AA, Alka O, Allard P-M, Barsch A, Cachet X, Caraballo-Rodriguez AM, Da Silva RR, Dang T, Garg N, Gauglitz JM, Gurevich A, Isaac G, Jarmusch AK, Kameník Z, Bin KK, Kessler N, Koester I, Korf A, Le Gouellec A, Ludwig M, Martin HC, McCall L-I, McSayles J, Meyer SW, Mohimani H, Morsy M, Moyne O, Neumann S, Neuweger H, Nguyen NH, Nothias-Esposito M, Paolini J, Phelan VV, Pluskal T, Quinn RA, Rogers S, Shrestha B, Tripathi A, van der Hooft JJJ, Vargas F, Weldon KC, Witting M, Yang H, Zhang Z, Zubeil F, Kohlbacher O, Böcker S, Alexandrov T, Bandeira N, Wang M, Dorrestein PC. Feature-based molecular networking in the GNPS analysis environment. Nat Methods. 2020;17:905–8. https://doi.org/10.1038/s41592-020-0933-6.
Quinn RA, Nothias L-F, Vining O, Meehan M, Esquenazi E, Dorrestein PC. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol Sci. 2017;38:143–54. https://doi.org/10.1016/j.tips.2016.10.011.
Olivon F, Elie N, Grelier G, Roussi F, Litaudon M, Touboul D. MetGem software for the generation of molecular networks based on the t-SNE algorithm. Anal Chem. 2018;90:13900–8. https://doi.org/10.1021/acs.analchem.8b03099.
Elie N, Santerre C, Touboul D. Generation of a molecular network from electron ionization mass spectrometry data by combining MZmine2 and MetGem software. Anal Chem. 2019;91:11489–92. https://doi.org/10.1021/acs.analchem.9b02802.
Neto FC, Raftery D. Expanding urinary metabolite annotation through integrated mass spectral similarity networking. Anal Chem. 2021;93:12001–10. https://doi.org/10.1021/acs.analchem.1c02041.
Le Daré B, Ferron P-J, Allard P-M, Clément B, Morel I, Gicquel T. New insights into quetiapine metabolism using molecular networking. Sci Rep. 2020;10:19921. https://doi.org/10.1038/s41598-020-77106-x.
Hayes RN, Gross ML. [10] Collision-induced dissociation. 1990;237–263.
Martín-Sómer A, Yáñez M, Gaigeot M-P, Spezia R. Unimolecular fragmentation induced by low-energy collision: statistically or dynamically driven? J Phys Chem A. 2014;118:10882–93. https://doi.org/10.1021/jp5076059.
Zhou Z, Luo M, Chen X, Yin Y, Xiong X, Wang R, Zhu Z-J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat Commun. 2020;11:4334. https://doi.org/10.1038/s41467-020-18171-8.
Calabrese V, Schmitz-Afonso I, Prevost C, Afonso C, Elomri A. Molecular networking and collision cross section prediction for structural isomer and unknown compound identification in plant metabolomics: a case study applied to Zhanthoxylum heitzii extracts. Anal Bioanal Chem. 2022;414:4103–18. https://doi.org/10.1007/s00216-022-04059-7.
An N, Zhu Q-F, Wang Y-Z, Xiong C-F, Hu Y-N, Feng Y-Q. Integration of chemical derivatization and in-source fragmentation mass spectrometry for high-coverage profiling of submetabolomes. Anal Chem. 2021;93:11321–8. https://doi.org/10.1021/acs.analchem.1c02673.
Kato S, Shimizu N, Ogura Y, Otoki Y, Ito J, Sakaino M, Sano T, Kuwahara S, Takekoshi S, Imagi J, Nakagawa K. Structural analysis of lipid hydroperoxides using mass spectrometry with alkali metals. J Am Soc Mass Spectrom. 2021;32:2399–409. https://doi.org/10.1021/jasms.1c00039.
Gass DT, Quintero AV, Hatvany JB, Gallagher ES. Metal adduction in mass spectrometric analyses of carbohydrates and glycoconjugates. Mass Spectrom Rev. 2022. https://doi.org/10.1002/mas.21801.
Poyer S, Laboureur L, Hebra T, Elie N, Van der Rest G, Salpin J-Y, Champy P, Touboul D. Dereplication of acetogenins from Annona muricata by combining tandem mass spectrometry after lithium and copper postcolumn cationization and molecular networks. J Am Soc Mass Spectrom. 2022;33:627–34. https://doi.org/10.1021/jasms.1c00303.
Qi Y, Volmer DA. Structural analysis of small to medium-sized molecules by mass spectrometry after electron-ion fragmentation (ExD) reactions. Analyst. 2016;141:794–806. https://doi.org/10.1039/C5AN02171E.
Lange M, Fedorova M. Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma. Anal Bioanal Chem. 2020;412:3573–84. https://doi.org/10.1007/s00216-020-02576-x.
Tang D, Zou L, Yin X, Ong CN. HILIC-MS for metabolomics: an attractive and complementary approach to RPLC-MS. Mass Spectrom Rev. 2016;35:574–600. https://doi.org/10.1002/mas.21445.
Laboureur L, Ollero M, Touboul D. Lipidomics by supercritical fluid chromatography. Int J Mol Sci. 2015;16:13868–84. https://doi.org/10.3390/ijms160613868.
van de Velde B, Guillarme D, Kohler I. Supercritical fluid chromatography – Mass spectrometry in metabolomics: past, present, and future perspectives. J Chromatogr B. 2020;1161:122444. https://doi.org/10.1016/j.jchromb.2020.122444.
Cody RB, Freiser BS. Electron impact excitation of ions from organics: an alternative to collision induced dissociation. Anal Chem. 1979;51:547–51. https://doi.org/10.1021/ac50040a022.
Cody RB, Freiser BS. Electron impact excitation of ions in Fourier transform mass spectrometry. Anal Chem. 1987;59:1054–6. https://doi.org/10.1021/ac00134a026.
Baba T, Ryumin P, Duchoslav E, Chen K, Chelur A, Loyd B, Chernushevich I. Dissociation of biomolecules by an intense low-energy electron beam in a high sensitivity time-of-flight mass spectrometer. J Am Soc Mass Spectrom. 2021;32:1964–75. https://doi.org/10.1021/jasms.0c00425.
Campbell JL, Baba T. Near-complete structural characterization of phosphatidylcholines using electron impact excitation of ions from organics. Anal Chem. 2015;87:5837–45. https://doi.org/10.1021/acs.analchem.5b01460.
Baba T, Campbell JL, Le Blanc JCY, Baker PRS. In-depth sphingomyelin characterization using electron impact excitation of ions from organics and mass spectrometry. J Lipid Res. 2016;57:858–67. https://doi.org/10.1194/jlr.M067199.
Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Structural identification of triacylglycerol isomers using electron impact excitation of ions from organics (EIEIO). J Lipid Res. 2016;57:2015–27. https://doi.org/10.1194/jlr.M070177.
Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Structural identification of eicosanoids with ring structures using differential mobility spectrometry–electron impact excitation of ions from organics mass spectrometry. J Am Soc Mass Spectrom. 2023;34:75–81. https://doi.org/10.1021/jasms.2c00256.
Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Distinguishing cis and trans isomers in intact complex lipids using electron impact excitation of ions from organics mass spectrometry. Anal Chem. 2017;89:7307–15. https://doi.org/10.1021/acs.analchem.6b04734.
Ducati AO, Ruskic D, Sosnowski P, Baba T, Bonner R, Hopfgartner G. Improved metabolite characterization by liquid chromatography – tandem mass spectrometry through electron impact type fragments from adduct ions. Anal Chim Acta. 2021;1150:338207. https://doi.org/10.1016/j.aca.2021.338207.
Che P, Davidson JT, Kool J, Kohler I. Electron activated dissociation - a complementary fragmentation technique to collision-induced dissociation for metabolite identification of synthetic cathinone positional isomers. Anal Chim Acta. 2023;1283:341962. https://doi.org/10.1016/j.aca.2023.341962.
Marzullo BP, Morgan TE, Wootton CA, Li M, Perry SJ, Saeed M, Barrow MP, O’Connor PB. Comparison of fragmentation techniques for the structural characterization of singly charged agrochemicals. Anal Chem. 2020;92:3143–51. https://doi.org/10.1021/acs.analchem.9b04820.
Weiss M, Macher JN, Seefeldt MA, Leese F. Molecular evidence for further overlooked species within the Gammarus fossarum complex (Crustacea: Amphipoda). Hydrobiologia. 2014;721:165–84. https://doi.org/10.1007/s10750-013-1658-7.
Chaumot A, Geffard O, Armengaud J, Maltby L. Gammarids as reference species for freshwater monitoring. In: Aquatic ecotoxicology. Elsevier, 2015;253–280.
Calabrese V, Salvador A, Clément Y, Brunet TA, Espeyte A, Chaumot A, Geffard O, Degli-Esposti D, Ayciriex S. Challenges and perspectives in MS-based omics approaches for ecotoxicology studies: an insight on Gammarids sentinel amphipods. Front Anal Sci. 2023;3.
Trapp J, Geffard O, Imbert G, Gaillard JC, Davin AH, Chaumot A, Armengaud J. Proteogenomics of gammarus fossarum to document the reproductive system of amphipods. Mol Cell Proteomics. 2014;13:3612–25. https://doi.org/10.1074/mcp.M114.038851.
Trapp J, Armengaud J, Pible O, Gaillard JC, Abbaci K, Habtoul Y, Chaumot A, Geffard O. Proteomic investigation of male Gammarus fossarum, a freshwater crustacean, in response to endocrine disruptors. J Proteome Res. 2015;14:292–303. https://doi.org/10.1021/pr500984z.
Leprêtre M, Geffard O, Espeyte A, Faugere J, Ayciriex S, Salvador A, Delorme N, Chaumot A, Degli-Esposti D. Multiple reaction monitoring mass spectrometry for the discovery of environmentally modulated proteins in an aquatic invertebrate sentinel species. Gammarus Fossarum Environ Pollut. 2022;315:120393. https://doi.org/10.1016/j.envpol.2022.120393.
Faugere J, Gouveia D, Ayciriex S, Chaumot A, Almunia C, François A, Armengaud J, Lemoine J, Geffard O, Degli-Esposti D, Salvador A. High-multiplexed monitoring of protein biomarkers in the sentinel Gammarus fossarum by targeted scout-MRM assay, a new vision for ecotoxicoproteomics. J Proteomics. 2020;226:103901. https://doi.org/10.1016/j.jprot.2020.103901.
Gouveia D, Chaumot A, Charnot A, Almunia C, François A, Navarro L, Armengaud J, Salvador A, Geffard O. Ecotoxico-proteomics for aquatic environmental monitoring: first in situ application of a new proteomics-based multibiomarker assay using caged amphipods. Environ Sci Technol. 2017;51:13417–26. https://doi.org/10.1021/acs.est.7b03736.
Koenig N, Almunia C, Bonnal-Conduzorgues A, Armengaud J, Chaumot A, Geffard O, Esposti DD. Co-expression network analysis identifies novel molecular pathways associated with cadmium and pyriproxyfen testicular toxicity in Gammarus fossarum. Aquat Toxicol. 2021;235:105816. https://doi.org/10.1016/j.aquatox.2021.105816.
Cosio C, Degli-Esposti D, Almunia C, Gaillet V, Sartelet H, Armengaud J, Chaumot A, Geffard O, Geffard A. Subcellular distribution of dietary methyl-mercury in Gammarus fossarum and its impact on the amphipod proteome. Environ Sci Technol. 2021;55:10514–23. https://doi.org/10.1021/acs.est.1c02385.
Gismondi E, Thomé J-P, Urien N, Uher E, Baiwir D, Mazzucchelli G, De Pauw E, Fechner LC, Lebrun JD. Ecotoxicoproteomic assessment of the functional alterations caused by chronic metallic exposures in gammarids. Environ Pollut. 2017;225:428–38. https://doi.org/10.1016/j.envpol.2017.03.006.
Fu T, Knittelfelder O, Geffard O, Clément Y, Testet E, Elie N, Touboul D, Abbaci K, Shevchenko A, Lemoine J, Chaumot A, Salvador A, Degli-Esposti D, Ayciriex S. Shotgun lipidomics and mass spectrometry imaging unveil diversity and dynamics in Gammarus fossarum lipid composition. iScience. 2021;24:102115. https://doi.org/10.1016/j.isci.2021.102115.
Fu T, Oetjen J, Chapelle M, Verdu A, Szesny M, Chaumot A, Degli-Esposti D, Geffard O, Clément Y, Salvador A, Ayciriex S. In situ isobaric lipid mapping by MALDI–ion mobility separation–mass spectrometry imaging. J Mass Spectrom. 2020;55. https://doi.org/10.1002/jms.4531.
Thomas Alexandre Brunet, Yohann Clément, Valentina Calabrese, Jérôme Lemoine, Olivier Geffard, Arnaud Chaumot, Davide Degli-Esposti, Arnaud Salvador, Sophie Ayciriex. Concomitant investigation of crustacean amphipods lipidome and metabolome during molting stage by Zeno SWATH Data-Independent Acquisition coupled with Electron Activated Dissociation and machine learning. Biorxiv Preprint. 2023.
Faugere J, Brunet TA, Clément Y, Espeyte A, Geffard O, Lemoine J, Chaumot A, Degli-Esposti D, Ayciriex S, Salvador A. Development of a multi-omics extraction method for ecotoxicology: investigation of the reproductive cycle of Gammarus fossarum. Talanta. 2023;253:123806. https://doi.org/10.1016/j.talanta.2022.123806.
Negri P, Krotulski A. Enhancing high-resolution mass spectrometry performance for NPS analysis with improved sensitivity and characterization. Toxicol Anal Clin. 2022;34:S149. https://doi.org/10.1016/j.toxac.2022.06.251.
Katajamaa M, Miettinen J, Orešič M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–6. https://doi.org/10.1093/bioinformatics/btk039.
Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. https://doi.org/10.1186/1471-2105-11-395.
Schmid R, Heuckeroth S, Korf A, Smirnov A, Myers O, Dyrlund TS, Bushuiev R, Murray KJ, Hoffmann N, Lu M, Sarvepalli A, Zhang Z, Fleischauer M, Dührkop K, Wesner M, Hoogstra SJ, Rudt E, Mokshyna O, Brungs C, Ponomarov K, Mutabdžija L, Damiani T, Pudney CJ, Earll M, Helmer PO, Fallon TR, Schulze T, Rivas-Ubach A, Bilbao A, Richter H, Nothias L-F, Wang M, Orešič M, Weng J-K, Böcker S, Jeibmann A, Hayen H, Karst U, Dorrestein PC, Petras D, Du X, Pluskal T. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat Biotechnol. 2023;41:447–9. https://doi.org/10.1038/s41587-023-01690-2.
Dührkop K, Fleischauer M, Ludwig M, Aksenov AA, Melnik AV, Meusel M, Dorrestein PC, Rousu J, Böcker S. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods. 2019;16:299–302. https://doi.org/10.1038/s41592-019-0344-8.
Böcker S, Letzel MC, Lipták Z, Pervukhin A. SIRIUS: decomposing isotope patterns for metabolite identification. Bioinformatics. 2009;25:218–24. https://doi.org/10.1093/bioinformatics/btn603.
Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci. 2015;112:12580–5. https://doi.org/10.1073/pnas.1509788112.
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–21. https://doi.org/10.1007/s11306-007-0082-2.
Lester R, Carey MC, Little JM, Cooperatein LA, Dowd SR. Crustacean intestinal detergent promotes sterol solubilization. Science (1979). 1975;189:1098–100. https://doi.org/10.1126/science.1162360.
Holwerda DA, Vonk HJ. Emulsifiers in the intestinal juice of crustacea. Isolation and nature of surface-active substances from Astacus leptodactylus Esch. and Homarus vulgaris L. Comp Biochem Physiol Part B Comp Biochem. 1973;45:51–8. https://doi.org/10.1016/0305-0491(73)90283-6.
Yamashita Y, Yamashita M. Identification of a novel selenium-containing compound, selenoneine, as the predominant chemical form of organic selenium in the blood of bluefin tuna. J Biol Chem. 2010;285:18134–8. https://doi.org/10.1074/jbc.C110.106377.
Belzile N, Chen Y-W, Gunn JM, Tong J, Alarie Y, Delonchamp T, Lang C-Y. The effect of selenium on mercury assimilation by freshwater organisms. Can J Fish Aquat Sci. 2006;63:1–10. https://doi.org/10.1139/f05-202.
Walter Helbling E, Fernando Menchi C, Villafañe VE. Bioaccumulation and role of UV-absorbing compounds in two marine crustacean species from Patagonia, Argentina. Photochem Photobiol Sci. 2002;1:820–5. https://doi.org/10.1039/b206584c.
Gouveia D, Pible O, Culotta K, Jouffret V, Geffard O, Chaumot A, Degli-Esposti D, Armengaud J. Combining proteogenomics and metaproteomics for deep taxonomic and functional characterization of microbiomes from a non-sequenced host. NPJ Biofilms Microbiomes. 2020;6. https://doi.org/10.1038/s41522-020-0133-2.
Godchaux W, Leadbetter ER. Sulfonolipids of gliding bacteria. Structure of the N-acylaminosulfonates. J Biol Chem. 1984;259:2982–90. https://doi.org/10.1016/S0021-9258(17)43246-7.
Walker A, Pfitzner B, Harir M, Schaubeck M, Calasan J, Heinzmann SS, Turaev D, Rattei T, Endesfelder D, Castell zu W, Haller D, Schmid M, Hartmann A, Schmitt-Kopplin P. Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci Rep. 2017;7:11047. https://doi.org/10.1038/s41598-017-10369-z.
Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, King N. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. Elife. 2012;1. https://doi.org/10.7554/eLife.00013.
Cordell GA, Lamahewage SNS. Ergothioneine, ovothiol A, and selenoneine—histidine-derived, biologically significant, trace global alkaloids. Molecules. 2022;27:2673. https://doi.org/10.3390/molecules27092673.
Christophersen C, Anthoni U. Organic sulfur compounds from marine organisms. Sulfur Rep. 1986;4:365–442. https://doi.org/10.1080/01961778608082487.
Rosenstock HM, Wallenstein MB, Wahrhaftig AL, Eyring H. Absolute rate theory for isolated systems and the mass spectra of polyatomic molecules. Proc Natl Acad Sci. 1952;38:667–78. https://doi.org/10.1073/pnas.38.8.667.
Fu T, Oetjen J, Chapelle M, Verdu A, Szesny M, Chaumot A, Degli‐Esposti D, Geffard O, Clément Y, Salvador A, Ayciriex S. In situ isobaric lipid mapping by MALDI–ion mobility separation–mass spectrometry imaging. J Mass Spectrom. 2020;55. https://doi.org/10.1002/jms.4531.
Funding
This work was supported by the French National Research Agency (ANR) (young investigator grant, ANR-18-CE34-0008 PLAN-TOX). VC was supported by a post-doctoral fellowship of the SENS research funding of the Université Claude Bernard Lyon 1. This work was performed within the framework of the EUR H2O’Lyon (ANR-17-EURE-0018) of Université de Lyon (UdL), within the program “Investissements d’Avenir” operated by the French National Research Agency (ANR). We thank the French GDR “Aquatic Ecotoxicology” framework which aims at fostering stimulating discussions and collaborations for more integrative approaches.
Author information
Authors and Affiliations
Contributions
VC: conceptualization; methodology; formal analysis; investigation; data analysis; visualization; writing—original draft. TAB: methodology, investigation. DDE: writing—review and editing. AC, OG: resources; writing—review and editing. AS: writing—review and editing. YC: resources; writing—review and editing. SA: conceptualization; funding acquisition; supervision; writing—original draft; writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ABC Highlights: authored by Rising Stars and Top Experts.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Calabrese, V., Brunet, T.A., Degli-Esposti, D. et al. Electron-activated dissociation (EAD) for the complementary annotation of metabolites and lipids through data-dependent acquisition analysis and feature-based molecular networking, applied to the sentinel amphipod Gammarus fossarum. Anal Bioanal Chem 416, 2893–2911 (2024). https://doi.org/10.1007/s00216-024-05232-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-024-05232-w