Abstract
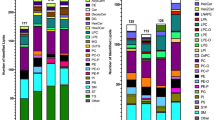
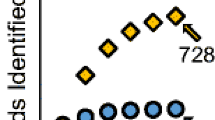
Untargeted lipidomics, with its ability to take a snapshot of the lipidome landscape, is an important tool to highlight lipid changes in pathology or drug treatment models. One of the shortcomings of most untargeted lipidomics based on UHPLC-HRMS is the low throughput, which is not compatible with large-scale screening. In this contribution, we evaluate the application of a sub-5-min high-throughput four-dimensional trapped ion mobility mass spectrometry (HT-4D-TIMS) platform for the fast profiling of multiple complex biological matrices. Human AC-16 cells and mouse brain, liver, sclera, and feces were used as samples. By using a fast 4-min RP gradient, the implementation of TIMS allows us to differentiate coeluting isomeric and isobaric lipids, with correct precursor ion isolation, avoiding co-fragmentation and chimeric MS/MS spectra. Globally, the HT-4D-TIMS allowed us to annotate 1910 different lipid species, 1308 at the molecular level and 602 at the sum composition level, covering 58 lipid subclasses, together with quantitation capability covering more than three orders of magnitude. Notably, TIMS values were highly comparable with respect to longer LC gradients (CV% = 0.39%). These results highlight how HT-4D-TIMS-based untargeted lipidomics possess high coverage and accuracy, halving the analysis time with respect to conventional UHPLC methods, and can be used for fast and accurate untargeted analysis of complex matrices to rapidly evaluate changes of lipid metabolism in disease models or drug discovery campaigns.
Graphical Abstract







Similar content being viewed by others
Abbreviations
- AHexCer:
-
Acylhexosylceramide
- ASM:
-
Acylsphingomyelin
- BMP:
-
Bismonoacylglycerophosphate
- CL:
-
Cardiolipin
- CAR:
-
Carnitine
- Cer:
-
Ceramide
- CE:
-
Cholesteryl ester
- CoQ:
-
Coenzyme Q
- DG:
-
Diacylglycerol
- DGGA:
-
Diacylglyceryl glucuronide
- DGDG:
-
Digalactosyldiacylglycerol
- Hex2Cer:
-
Dihexosylceramide
- DCAE:
-
Esterified deoxycholic acid
- DG O-:
-
Ether-linked diacylglycerol
- LPC O-:
-
Ether-linked lysophosphatidylcholine
- LPE O-:
-
Ether-linked lysophosphatidylethanolamine
- MGDG O-:
-
Ether-linked monogalactosyldiacylglycerol
- Ox-PE-O-:
-
Ether-linked oxidized phosphatidylethanolamine
- PC O-:
-
Ether-linked phosphatidylcholine
- PE O-:
-
Ether-linked phosphatidylethanolamine
- PG O-:
-
Ether-linked phosphatidylglycerol
- PI O-:
-
Ether-linked phosphatidylinositol
- PS O-:
-
Ether-linked phosphatidylserine
- TG O-:
-
Ether-linked triacylglycerol
- FA:
-
Fatty acid
- FAHFA:
-
Fatty acid ester of hydroxyl fatty acid
- GM3:
-
Ganglioside GM3
- HBMP:
-
Hemibismonoacylglycerophosphate
- HexCer:
-
Hexosylceramide
- LPC:
-
Lysophophatidylcholine
- LPE:
-
Lysophosphatidylethanolamine
- LPG:
-
Lysophosphatidylglycerol
- LPI:
-
Lysophosphatidylinositol
- LPS:
-
Lysophosphatidylserine
- MG:
-
Monoacylglycerol
- MGDG:
-
Monogalactosyldiacylglycerol
- NAGly:
-
N-Acyl glycine
- NAGlySer:
-
N-Acyl glycyl serine
- Ox-PI-Cer:
-
Oxidized ceramide phosphoinositol
- Ox-PE:
-
Oxidized phosphatidylethanolamine
- Ox-PI:
-
Oxidized phosphatidylinositol
- Ox-SM:
-
Oxidized sphingomyelin
- Ox-SHexCer:
-
Oxidized sulfatide
- PC:
-
Phosphatidylcholine
- PEtOH:
-
Phosphatidylethanol
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- PI:
-
Phosphatidylinositol
- PMeOH:
-
Phosphatidylmethanol
- PS:
-
Phosphatidylserine
- SPB:
-
Sphingoid bases
- SM:
-
Sphingomyelin
- ST:
-
Sterol
- SE:
-
Sterol ester
- SHexCer:
-
Sulfatide
- TG:
-
Triacylglycerol
- Hex3Cer:
-
Trihexosylceramide
- Vit. D:
-
Vitamin D
References
Wenk M. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. https://doi.org/10.1038/nrd1776.
Han X, Gross RW. The foundations and development of lipidomics. J Lipid Res. 2022;63(2): 100164. https://doi.org/10.1016/j.jlr.2021.100164.
Züllig T, Trötzmüller M, Köfeler HC. Lipidomics from sample preparation to data analysis: a primer. Anal Bioanal Chem. 2020;412:2191–209. https://doi.org/10.1007/s00216-019-02241-y.
Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt Chem. 2014;61:192–206. https://doi.org/10.1016/j.trac.2014.04.017.
Xuan Q, Hu C, Yu D, Wang L, Zhou Y, Zhao X, Li Q, Hou X, Xu G. Development of a high coverage pseudotargeted lipidomics method based on ultra-high performance liquid chromatography-mass spectrometry. Anal Chem. 2018;90(12):7608–16. https://doi.org/10.1021/acs.analchem.8b01331.
Xu T, Hu C, Xuan Q, Xu G. Recent advances in analytical strategies for mass spectrometry-based lipidomics. Anal Chim Acta. 2020;1137:156–69. https://doi.org/10.1016/j.aca.2020.09.060.
Kostidis S, Sánchez-López E, Giera M. Lipidomics analysis in drug discovery and development. Curr Opin Chem Biol. 2023;72: 102256. https://doi.org/10.1016/j.cbpa.2022.102256.
Zhang Y, Xie Y, Lv W, Hu C, Xu T, Liu X, Zhang R, Xu G, Xia Y, Zhao X. A high throughput lipidomics method and its application in atrial fibrillation based on 96-well plate pretreatment and liquid chromatography-mass spectrometry. J Chromatogr A. 2021;1651: 462271. https://doi.org/10.1016/j.chroma.2021.462271.
Jung HR, Sylvänne T, Koistinen KM, Tarasov K, Kauhanen D, Ekroos K. High throughput quantitative molecular lipidomics. Biochim Biophys Acta. 2011;1811(11):925–34. https://doi.org/10.1016/j.bbalip.2011.06.025.
Paglia G, Kliman M, Claude E, Scoot G, Astarita G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal Bioanal Chem. 2015;407:4995–5007. https://doi.org/10.1007/s00216-015-8664-8.
Hinz C, Liggi S, Griffin JL. The potential of ion mobility mass spectrometry for high-throughput and high-resolution lipidomics. Curr Opin Chem Biol. 2018;42:42–50. https://doi.org/10.1016/j.cbpa.2017.10.018.
Vasilopoulou CG, Sulek K, Brunner AD, Meitei N, Schweiger-Hufnagel U, Meyer SW, Barsch A, Mann M, Meier F. Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts. Nat Commun. 2020;11:331. https://doi.org/10.1038/s41467-019-14044-x.
Meier F, Park MA, Mann M. Trapped ion mobility spectrometry and parallel accumulation-serial fragmentation in proteomics. Mol Cell Proteomics. 2021;20: 100138. https://doi.org/10.1016/j.mcpro.2021.100138.
Merciai F, Musella S, Sommella E, Bertamino A, D’Ursi AM, Campiglia P. Development and application of a fast ultra-high performance liquid chromatography-trapped ion mobility mass spectrometry method for untargeted lipidomics. J Chromatogr A. 2022;1673: 463124. https://doi.org/10.1016/j.chroma.2022.463124.
Skowronek P, Thielert M, Voytik E, Tanzer MC, Hansen FM, Willems S, Karayel O, Brunner AD, Meier F, Mann M. Rapid and in-depth coverage of the (phospho-)proteome with deep libraries and optimal window design for dia-PASEF. Mol Cell Proteomics. 2022;21(9): 100279. https://doi.org/10.1016/j.mcpro.2022.100279.
Chen X, Yin Y, Luo M, Zhou Z, Cai Y, Zhu ZJ. Trapped ion mobility spectrometry-mass spectrometry improves the coverage and accuracy of four-dimensional untargeted lipidomics. Anal Chim Acta. 2022;1210: 339886. https://doi.org/10.1016/j.aca.2022.339886.
Lerner R, Baker D, Schwitter C, Neuhaus S, Hauptmann T, Post JM, Kramer S, Bindila L. Four-dimensional trapped ion mobility spectrometry lipidomics for high throughput clinical profiling of human blood samples. Nat Commun. 2023;14(1):937. https://doi.org/10.1038/s41467-023-36520-1.
Ciccarelli M, Merciai F, Carrizzo A, Sommella E, Di Pietro P, Caponigro V, Salviati E, Musella S, Sarno VD, Rusciano M, Toni AL, Iesu P, Izzo C, Schettino G, Conti V, Venturini E, Vitale C, Scarpati G, Bonadies D, Rispoli A, Polverino B, Poto S, Pagliano P, Piazza O, Licastro D, Vecchione C, Campiglia P. Untargeted lipidomics reveals specific lipid profiles in COVID-19 patients with different severity from Campania region (Italy). J Pharm Biomed Anal. 2022;217: 114827. https://doi.org/10.1016/j.jpba.2022.114827.
Tumanov S, Kamphorst JJ. Recent advances in expanding the coverage of the lipidome. Curr Opin Biotechnol. 2017;43:127–33. https://doi.org/10.1016/j.copbio.2016.11.008.
Zhang F, Guo S, Zhang M, Zhang Z, Guo Y. Characterizing ion mobility and collision cross section of fatty acids using electrospray ion mobility mass spectrometry. J Mass Spectrom. 2015;50(7):906–13. https://doi.org/10.1002/jms.3600.
Xiang H, Zhang B, Wang Y, Xu N, Zhang F, Luo R, Ji M, Ding C. Region-resolved multi-omics of the mouse eye. Cell Rep. 2023;42(2): 112121. https://doi.org/10.1016/j.celrep.2023.112121.
Rust BM, Picklo MJ, Yan L, Mehus AA, Zeng H. Time-restricted feeding modifies the fecal lipidome and the gut microbiota. Nutrients. 2023;15(7):1562. https://doi.org/10.3390/nu15071562.
dkowiak J, Jirásko R, Kolářová D, Bártl J, Hájek T, Antonelli M, Vaňková Z, Wolrab D, Hrstka R, Študentová H, Melichar B, Pešková K, Holčapek M. Robust and high-throughput lipidomic quantitation of human blood samples using flow injection analysis with tandem mass spectrometry for clinical use. Anal Bioanal Chem. 2023;415(5):935-951. https://doi.org/10.1007/s00216-022-04490-w.
Cajka T, Smilowitz JT, Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal Chem. 2017;89(22):12360–8. https://doi.org/10.1021/acs.analchem.7b03404.
Koelmel JP, Cochran JA, Ulmer CZ, Levy AJ, Patterson RE, Olsen BC, Yost RA, Bowden JA, Garrett TJ. Software tool for internal standard based normalization of lipids, and effect of data-processing strategies on resulting values. BMC Bioinformatics.2019;20(1): 217. https://doi.org/10.1186/s12859-019-2803-8.
Bartosova Z, Gonzalez SV, Voigt A, Bruheim P. High throughput semiquantitative UHPSFC-MS/MS lipid profiling and lipid class determination. J Chromatogr Sci. 2021;59(7):670–80. https://doi.org/10.1093/chromsci/bmaa121.
Drotleff B, Illison J, Schlotterbeck J, Lukowski R, Lämmerhofer M. Comprehensive lipidomics of mouse plasma using class-specific surrogate calibrants and SWATH acquisition for large-scale lipid quantification in untargeted analysis. Anal Chim Acta. 2019;1086:90–102. https://doi.org/10.1016/j.aca.2019.08.030.
Choi J, Yin T, Shinozaki K, Lampe JW, Stevens JF, Becker LB, Kim J. Comprehensive analysis of phospholipids in the brain, heart, kidney, and liver: brain phospholipids are least enriched with polyunsaturated fatty acids. Mol Cell Biochem. 2018;442(1–2):187–201. https://doi.org/10.1007/s11010-017-3203-x.
Acknowledgements
Cells and tissues were kindly donated as part of secondary experiments from Prof. Carrizzo the Department of Medicine (DIPMED, University of Salerno, Italy) and from Prof. Paolisso of the Department of Advanced Medical and Surgical Sciences, University of Campania “Luigi Vanvitelli”, Naples, Italy.
Funding
This work was supported by Ministero dell’Università e della Ricerca (MIUR) project PIR01_00032 BIO OPEN LAB BOL “CUP” J37E19000050007, project CIR01_00032 – BOL “BIO Open Lab—Rafforzamento del capitale umano” and project PNC0000001 D34 Health—Digital Driven Diagnostics, prognostics and therapeutics for sustainable Health care “CUP” B53C22006090001 and Regione Campania (Italy) grant “Combattere la resistenza tumorale: piattaforma integrata multi-disciplinare per un approccio tecnologico innovative alle oncoterapie—CAMPANIA ONCO-TERAPIE” project number B61G18000470007. to P. Campiglia.
Author information
Authors and Affiliations
Contributions
Conceptualization: FM. Methodology: VC. Formal analysis and investigation: DLG, ES. Data curation: TC. Writing—original draft preparation: MGB. Writing—review and editing: ES, SM. Funding acquisition: PC. Supervision: ES, CC.
Corresponding author
Ethics declarations
Ethics approval
The study was conducted in conformity with Italian (D.L. 26/2014) and European (directive 2010/63/EU) regulations on the protection of animals used for scientific purposes and approved by the Italian Ministry.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Merciai, F., Basilicata, M.G., La Gioia, D. et al. Sub-5-min RP-UHPLC-TIMS for high-throughput untargeted lipidomics and its application to multiple matrices. Anal Bioanal Chem 416, 959–970 (2024). https://doi.org/10.1007/s00216-023-05084-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-023-05084-w