Abstract
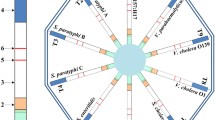
A simple and rapid polymerase chain reaction (PCR)-based lateral flow assay (LFA) was developed for multiplex detection of hygiene indicator bacteria. Specifically, new PCR primers were designed for accurately detecting Escherichia coli, coliform bacteria, and total bacteria, and the results obtained as a colorimetric signal (generated by the accumulation of gold nanoparticles at distinct test zones on flow strips) could be identified by the naked eye in <10 min after the completion of PCR. The proposed LFA system did not exhibit any cross-reactivities with 8 distinct bacterial strains and can detect down to 1 colony forming unit (CFU)/mL. Furthermore, three species of cultured bacteria (Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa) inoculated onto sterilized ham were successfully analyzed using the LFA system, which demonstrated that this system shows sufficient sensitivity and specificity for food hygiene monitoring. The speed and simplicity of this LFA make it suitable for use in the food industry as part of routine screening analysis.





Similar content being viewed by others
References
World Health Organization (WHO). WHO estimates of the global burden of foodborne diseases: Food-Borne Disease Burden Epidemiology Reference Group. 2015.
Del Portal DA, Karras DJ. Surveillance for foodborne disease outbreaks - United States, 2009-2010. Ann Emerg Med. 2013;62:91–3. https://doi.org/10.1016/j.annemergmed.2013.04.001.
Davidson PM, Roth LA, Gambrel-Lenarz SA. Chapter 7 coliform and other indicator bacteria. In: Standard methods for the examination of dairy products. American Public Health Association. 2004.
Morton RD. Aerobic Plate Count. In: R, editor. Compendium of methods for the microbiological examination of foods, 8th ed. Gaithersburg: Food and Drug Administration; 2001.
Moore G, Griffith C. A comparison of traditional and recently developed methods for monitoring surface hygiene within the food industry: an industry trial. Int J Environ Health Res. 2002;12:317–29. https://doi.org/10.1080/0960312021000056429.
Edberg SC, Allen MJ, Smith DB, LeChevallier M, Kriz N, Callan D, et al. National field evaluation of a defined substrate method for the simultaneous detection of total coliforms and Escherichia coli from drinking water: comparison with presence-absence techniques. Appl Environ Microbiol. 1989;55:1003–8. https://doi.org/10.1128/aem.55.4.1003-1008.1989.
Rompré A, Servais P, Baudart J, De-Roubin MR, Laurent P. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J Microbiol Methods. 2002;49:31–54. https://doi.org/10.1016/S0167-7012(01)00351-7.
Klein H, Fung DYC. Identification and quantification of fecal coliforms using violet red bile agar at elevated temperature. J Milk Food Technol. 1976;39:768–70. https://doi.org/10.4315/0022-2747-39.11.768.
Hartman PA. Further studies on the selectivity of violet red bile agar. J Milk Food Technol. 1960;23:45–8. https://doi.org/10.4315/0022-2747-23.2.45.
ISO-4832. ISO 4831:2006 Microbiology of food and animal feeding stuffs — horizontal method for the detection and enumeration of coliforms — most probable number technique. ICS 0710030 Food Microbiol. 2006;3:11
Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–50. https://doi.org/10.1016/0076-6879(87)55023-6.
Bej AK, McCarty SC, Atlas RM. Detection of coliform bacteria and Escherichia coli by multiplex polymerase chain reaction: comparison with defined substrate and plating methods for water quality monitoring. Appl Environ Microbiol. 1991;57:2429–32. https://doi.org/10.1128/aem.57.8.2429-2432.1991.
Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–94. https://doi.org/10.1101/gr.6.10.986.
Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–66. https://doi.org/10.1099/00221287-148-1-257.
Mens PF, van Amerongen A, Sawa P, Kager PA, Schallig HDFH. Molecular diagnosis of malaria in the field: development of a novel 1-step nucleic acid lateral flow immunoassay for the detection of all 4 human Plasmodium spp. and its evaluation in Mbita, Kenya. Diagn Microbiol Infect Dis. 2008;61:421–7. https://doi.org/10.1016/j.diagmicrobio.2008.03.009.
Ang GY, Yu CY, Yean CY. Ambient temperature detection of PCR amplicons with a novel sequence-specific nucleic acid lateral flow biosensor. Biosens Bioelectron. 2012;38:151–6. https://doi.org/10.1016/j.bios.2012.05.019.
Rastogi SK, Gibson CLM, Branen JR, Aston DE, Branen AL, Hrdlicka PJ. DNA detection on lateral flow test strips: enhanced signal sensitivity using LNA-conjugated gold nanoparticles. Chem Commun. 2012;48:7714–6. https://doi.org/10.1039/c2cc33430e.
Roskos K, Hickerson AI, Lu HW, Ferguson TM, Shinde DN, Klaue Y, et al. Simple system for isothermal DNA amplification coupled to lateral flow detection. PLoS One. 2013;8:e69355. https://doi.org/10.1371/journal.pone.0069355.
Li J, Macdonald J. Multiplexed lateral flow biosensors: technological advances for radically improving point-of-care diagnoses. Biosens Bioelectron. 2016;3:177–92.
Li J, Pollak NM, MacDonald J. Multiplex detection of nucleic acids using recombinase polymerase amplification and a molecular colorimetric 7-segment display. ACS Omega. 2019;4:11388–96. https://doi.org/10.1021/acsomega.9b01097.
Park BH, Oh SJ, Jung JH, Choi G, Seo JH, Kim DH, et al. An integrated rotary microfluidic system with DNA extraction, loop-mediated isothermal amplification, and lateral flow strip based detection for point-of-care pathogen diagnostics. Biosens Bioelectron. 2017;91:334–40. https://doi.org/10.1016/j.bios.2016.11.063.
Pan Q, Zhu J, Liu L, Cong Y, Hu F, Li J, et al. Functional identification of a putative β-galactosidase gene in the special lac gene cluster of lactobacillus acidophilus. Curr Microbiol. 2010;60:172–8. https://doi.org/10.1007/s00284-009-9521-9.
Maheux AF, Boudreau DK, Bisson MA, Dion-Dupont V, Bouchard S, Nkuranga M, et al. Molecular method for detection of total coliforms in drinking water samples. Appl Environ Microbiol. 2014;80:4074–84. https://doi.org/10.1128/AEM.00546-14.
Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip. 2016;16:753–63. https://doi.org/10.1039/c5lc01392e.
Kalogianni DP, Goura S, Aletras AJ, Christopoulos TK, Chanos MG, Christofidou M, et al. Dry reagent dipstick test combined with 23S rRNA PCR for molecular diagnosis of bacterial infection in arthroplasty. Anal Biochem. 2007;361:169–75. https://doi.org/10.1016/j.ab.2006.11.013.
Edwards KA, Baeumner AJ. Optimization of DNA-tagged dye-encapsulating liposomes for lateral-flow assays based on sandwich hybridization. Anal Bioanal Chem. 2006;386:1335–43. https://doi.org/10.1007/s00216-006-0705-x.
Baeumner AJ, Jones C, Wong CY, Price A. A generic sandwich-type biosensor with nanomolar detection limits. Anal Bioanal Chem. 2004;378:1587–93. https://doi.org/10.1007/s00216-003-2466-0.
Baeumner AJ, Pretz J, Fang S. A universal nucleic acid sequence biosensor with nanomolar detection limits. Anal Chem. 2004;76:888–94. https://doi.org/10.1021/ac034945l.
San Millán RM, Martínez-Ballesteros I, Rementeria A, Garaizar J, Bikandi J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res Notes. 2013;6:513. https://doi.org/10.1186/1756-0500-6-513.
Molina F, López-Acedo E, Tabla R, Roa I, Gómez A, Rebollo JE. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol. 2015;15:1–9. https://doi.org/10.1186/s12896-015-0168-2.
Naravaneni R, Jamil K. Rapid detection of food-borne pathogens by using molecular techniques. J Med Microbiol. 2005;54:51–4. https://doi.org/10.1099/jmm.0.45687-0.
Hoorfar J. Rapid detection, characterization, and enumeration of foodborne pathogens. APMIS. 2011;119:1–24. https://doi.org/10.1111/j.1600-0463.2011.02767.x.
Galikowska E, Kunikowska D, Tokarska-Pietrzak E, Dziadziuszko H, Łoś JM, Golec P, et al. Specific detection of Salmonella enterica and Escherichia coli strains by using ELISA with bacteriophages as recognition agents. Eur J Clin Microbiol Infect Dis. 2011;30:1067–73. https://doi.org/10.1007/s10096-011-1193-2.
Kuo JT, Cheng CY, Huang HH, Tsao CF, Chung YC. A rapid method for the detection of representative coliforms in water samples: polymerase chain reaction-enzyme-linked immunosorbent assay (PCR-ELISA). J Ind Microbiol Biotechnol. 2010;37:237–44. https://doi.org/10.1007/s10295-009-0666-0.
Availability of data and materials
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Code availability
Not applicable.
Funding
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korea Government (Ministry of Science and ICT) (NRF-2020R1C1C1012275) and from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Agro and Livestock Products Safety·Flow Management Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (IPET319112-2).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 796 kb)
Rights and permissions
About this article
Cite this article
Kim, J.M., Park, J.S., Yoon, T.H. et al. Nucleic acid lateral flow assay for simultaneous detection of hygiene indicator bacteria. Anal Bioanal Chem 413, 5003–5011 (2021). https://doi.org/10.1007/s00216-021-03462-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03462-w