Abstract
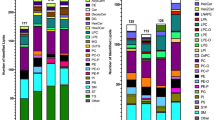
Pseudotargeted analysis combines the advantages of untargeted and targeted lipidomics methods based on chromatography-mass spectrometry (MS). This study proposed a comprehensive pseudotargeted lipidomics method based on three-phase liquid extraction (3PLE) and segment data-dependent acquisition (SDDA). We used a 3PLE method to extract the lipids with extensive coverage from biological matrixes. 3PLE was composed of one aqueous and two organic phases. The upper and middle organic phases enriched neutral lipids and glycerophospholipids, respectively, combined and detected together. Besides, the SDDA strategy improved the detection of co-elution ions in the lipidomics analysis. A total of 554 potential lipids were detected by the developed approach in both positive and negative modes using ultra-high-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS). Compared with the conventional liquid-liquid extraction (LLE) approaches, including methyl tert-butyl ether (MTBE) and Bligh-Dyer (BD) methods, 3PLE combined with SDDA significantly increased the lipid coverage 87.2% and 89.7%, respectively. Also, the proposed pseudotargeted lipidomics approach exhibited higher sensitivity and better repeatability than the untargeted approach. Finally, we applied the established pseudotargeted method to the plasma lipid profiling from the depressed rats and screened 61 differential variables. The results demonstrated that the pseudotargeted method based on 3PLE and SDDA broadened lipid coverage and improved the detection of co-elution ions with excellent sensitivity and precision, indicating significant potential for the lipidomics analysis.
Graphical abstract







Similar content being viewed by others
Abbreviations
- MS:
-
Mass spectrometry
- 3PLE:
-
Three-phase liquid extraction
- SDDA:
-
Segment data-dependent acquisition
- UHPLC-MS/MS:
-
Ultra-high-performance liquid chromatography-tandem mass spectrometry
- LLE:
-
Liquid-liquid extraction
- MTBE:
-
Methyl tert-butyl ether
- BD:
-
Bligh-Dyer
- PL:
-
Phospholipid
- LPL:
-
Lysophospholipid
- MRM:
-
Multiple reaction monitoring
- Q-TOF:
-
Quadrupole-time-of-flight
- TQ:
-
Triple quadrupole
- IS:
-
Internal standard
- LPE:
-
Lysophosphatidylethanolamine
- SM:
-
Sphingomyelin
- Cer:
-
Ceramide
- PC:
-
Phosphatidylcholine
- TG:
-
Triglyceride
- QC:
-
Quality control
- S/N:
-
Signal to noise
- RSD:
-
Relative standard deviation
- PLS-DA:
-
Partial least squares-discriminant analysis
- OPLS-DA:
-
Orthogonal partial least squares-discriminant analysis
- FDR:
-
False discovery rate
- HMDB:
-
Human metabolome database
- MG:
-
Monoglyceride
- DG:
-
Diglyceride
- LPC:
-
Lysophosphatidylcholine
- CE:
-
Cholesteryl ester
- LOD:
-
Limit of determination
- LOQ:
-
Limit of quantitation
- VIP:
-
Variable importance of projection
- AA:
-
Arachidonic acid
- FA:
-
Fatty acid
- PE:
-
Phosphatidylethanolamine
- PLA:
-
Phospholipase A
- LOX:
-
Lipoxygenase
- COX:
-
Cyclooxygenase
- HpETE:
-
Hydroperoxyeicosatetraenoic acid
- LT:
-
Leukotriene
- PG:
-
Prostaglandin
- TX:
-
Thromboxane
- SMase:
-
Sphingomyelinase
- CDase:
-
Ceramidase
- SP:
-
Sphingolipid
- GP:
-
Glycerophospholipid
- GL:
-
Glycerolipid
References
World Health Organization. Depression and other common mental disorders. 2017.
Kessler RC. The costs of depression. Psychiatr Clin N Am. 2012;35:1–14.
Ösby U, Brandt L, Correia N, Ekbom A, Sparén P. Excess mortality in bipolar and unipolar disorder in Sweden. Arch Gen Psychiatry. 2001;58:844–50.
Carvalho AF, Berk M, Hyphantis TN, McIntyre RS. The integrative management of treatment-resistant depression: a comprehensive review and perspectives. Psychother Psychosom. 2014;83:70–88.
Wang Y, Shi Y, Xu Z, Fu H, Zeng H, Zheng G. Efficacy and safety of Chinese herbal medicine for depression: a systematic review and meta-analysis of randomized controlled trials. J Psychiatr Res. 2019;117:74–91.
Wang W, Hu X, Zhao Z, Liu P, Hu Y, Zhou H, et al. Antidepressant-like effects of liquiritin and isoliquiritin from Glycyrrhiza uralensis in the forced swimming test and tail suspension test in mice. Prog Neuro-Psychopharmacol. 2008;32:1179–84.
Li M, Yang L, Bai Y, Liu H. Analytical methods in lipidomics and their applications. Anal Chem. 2014;86:161–75.
Teo CC, Chong WPK, Tan E, Basri NB, Low ZJ, Ho YS. Advances in sample preparation and analytical techniques for lipidomics study of clinical samples. Trac-Trend Anal Chem. 2015;66:1–18.
Brunkhorst-Kanaan N, Klatt-Schreiner K, Hackel J, Schröeter K, Trautmann S, Hahnefeld L, et al. Targeted lipidomics reveal derangement of ceramides in major depression and bipolar disorder. Metabolism. 2019;95:65–76.
Liu X, Li J, Zheng P, Zhao X, Zhou C, Hu C, et al. Plasma lipidomics reveals potential lipid markers of major depressive disorder. Anal Bioanal Chem. 2016;408:6497–507.
Xu T, Hu C, Xuan Q, Xu G. Recent advances in analytical strategies for mass spectrometry-based lipidomics. Anal Chim Acta. 2020;1137:156–69.
Luque de Castro MD, Quiles-Zafra R. Lipidomics: an omics discipline with a key role in nutrition. Talanta. 2020;219.
Yuan Z, Majchrzak-Hong S, Keyes GS, Iadarola MJ, Mannes AJ, Ramsden CE. Lipidomic profiling of targeted oxylipins with ultra-performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2018;410:6009–29.
Chao Y, Gao S, Wang X, Li N, Zhao H, Wen X, et al. Untargeted lipidomics based on UPLC-QTOF-MS/MS and structural characterization reveals dramatic compositional changes in serum and renal lipids in mice with glyoxylate-induced nephrolithiasis. J Chromatogr B. 2018;1095:258–66.
Li Y, Ruan Q, Li Y, Ye G, Lu X, Lin X, et al. A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring. J Chromatogr A. 2012;1255:228–36.
Xuan Q, Hu C, Yu D, Wang L, Zhou Y, Zhao X, et al. Development of a high coverage pseudotargeted lipidomics method based on ultra-high performance liquid chromatography-mass spectrometry. Anal Chem. 2018;90:7608–16.
Lv W, Wang L, Xuan Q, Zhao X, Liu X, Shi X, et al. Pseudotargeted method based on parallel column two- dimensional liquid chromatography-mass spectrometry for broad coverage of metabolome and lipidome. Anal Chem. 2020;92:6043–50.
Yang J, Liu D, Jin W, Zhong Q, Zhou T. A green and efficient pseudotargeted lipidomics method for the study of depression based on ultra-high performance supercritical fluid chromatography-tandem mass spectrometry. J Pharm Biomed. 2021.
Wang Y, Liu F, Li P, He C, Wang R, Su H, et al. An improved pseudotargeted metabolomics approach using multiple ion monitoring with time-staggered ion lists based on ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Anal Chim Acta. 2016;927:82–8.
Calderón-Santiago M, Priego-Capote F, Luque de Castro MD. Enhanced detection and identification in metabolomics by use of LC-MS/MS untargeted analysis in combination with gas-phase fractionation. Anal Chem. 2014;86:7558–65.
Yang J, Jin W, Liu D, Zhong Q, Zhou T. Enhanced pseudotargeted analysis using a segment data dependent acquisition strategy by liquid chromatography-tandem mass spectrometry for a metabolomics study of liquiritin in the treatment of depression. J Sep Sci. 2020;43:2088–96.
Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7.
Carlson LA. Extraction of lipids from human whole serum and lipoproteins and from rat liver tissue with methylene chloride-methanol: a comparison with extraction with chloroform-methanol. Clin Chim Acta. 1985;149:89–93.
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–46.
Vale G, Martin SA, Mitsche MA, Thompson BM, Eckert KM, McDonald JG. Three-phase liquid extraction: a simple and fast method for lipidomic workflows. J Lipid Res. 2019;60:694–706.
Shibusawa Y, Yamakawa Y, Noji R, Yanagida A, Shindo H, Ito Y. Three-phase solvent systems for comprehensive separation of a wide variety of compounds by high-speed counter-current chromatography. J Chromatogr A. 2006;1133:119–25.
Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50.
Demirkan A, Isaacs A, Ugocsai P, Liebisch G, Struchalin M, Rudan I, et al. Plasma phosphatidylcholine and sphingomyelin concentrations are associated with depression and anxiety symptoms in a Dutch family-based lipidomics study. J Psychiatr Res. 2013;47:357–62.
Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67.
Miranda AM, Oliveira TG. Lipids under stress - a lipidomic approach for the study of mood disorders. Bioessays. 2015;37:1226–35.
Castro-Perez JM, Kamphorst J, DeGroot J, Lafeber F, Goshawk J, Yu K, et al. Comprehensive LC-MSE lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J Proteome Res. 2010;9:2377–89.
Mocking RJT, Assies J, Ruhé HG, Schene AH. Focus on fatty acids in the neurometabolic pathophysiology of psychiatric disorders. J Inherit Metab Dis. 2018;41:597–611.
Furuyashiki T, Akiyama S, Kitaoka S. Roles of multiple lipid mediators in stress and depression. Int Immunol. 2019;31:579–87.
Berglund L, Sacks F, Brunzell JD. Risk factors for cardiovascular disease: renewed interest in triglycerides. Clin Lipidol. 2013;8:1–4.
Lee H, Kim Y. Serum lipid levels and suicide attempts. Acta Psychiatr Scand. 2003;108:215–21.
Jagenburg R, Svanborg A. Self-induced protein-calorie malnutrition in a healthy adult male. A study of plasma proteins, free amino acids and lipids. Acta Med Scand. 1968;183:67–71.
Qu Y, Chang L, Klaff J, Seemann R, Rapoport SI. Imaging brain phospholipase A2-mediated signal transduction in response to acute fluoxetine administration in unanesthetized rats. Neuropsychopharmacol. 2003;28:1219–26.
Funding
This work was supported by the National Natural Science Foundation of China (No. 21775047), and the Natural Science Foundation of Guangdong Province, China (No. 2021A1515012323).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Research involving human participants and/or animals
This animal study was approved by the Committee on the Ethics of Animal Experiments of Guangdong Pharmaceutical University (No.00199407). We executed animal experiments in accordance with Regulations on Animal Experiments in Guangdong Pharmaceutical University. We did not use samples from human in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 15.4 mb)
Rights and permissions
About this article
Cite this article
Liu, D., Yang, J., Jin, W. et al. A high coverage pseudotargeted lipidomics method based on three-phase liquid extraction and segment data-dependent acquisition using UHPLC-MS/MS with application to a study of depression rats. Anal Bioanal Chem 413, 3975–3986 (2021). https://doi.org/10.1007/s00216-021-03349-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03349-w