Abstract
Effective and simultaneous monitoring of the abnormal expression of certain microRNAs (miRNAs), especially for miRNA-21 and miRNA-155, can indicate drug resistance in lung cancer. In this work, T7 exonuclease (T7 Exo)-assisted target recycling amplification coupled with the extensive fluorescence quenching of graphene oxide (GO) was designed for the simultaneous detection of miRNA-21 and miRNA-155 using FAM- and ROX-labeled single-strand DNA probes. Through this method, the variable emission intensities of FAM and ROX caused by the introduction of miRNA-21 and miRNA-155, respectively, were obtained with high sensitivity. The method exhibited excellent analytical performance for simultaneous detection of miRNA-21 and miRNA-155 without cross-interference. The linear range was from 0.005 nM to 5 nM over three orders of magnitude, with detection limits as low as 3.2 pM and 4.5 pM for miRNA-21 and miRNA-155, respectively. Furthermore, the recovery (92.49–103.67%) and relative standard deviation (RSD < 4.8%) of the standard addition test of miRNA-21 and miRNA-155 in human plasma suggested the potential for drug resistance warning in clinical practice via this simple strategy.
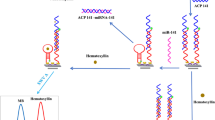
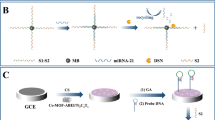
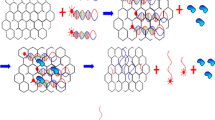
Graphical abstract

A homogeneous T7 Exo-assisted signal amplification combined with GO quenching platform was developed for accurate, sensitive and simultaneous analysis of miRNA-21 and miRNA-155 for drug resistance warning in lung cancer. This simple method exhibited a wide linear range and low LODs for miR-21 and miR-155.





Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics 2020. CA Cancer J Clin. 2020;67:7–30.
Dolly SO, Collins DC, Sundar R, Popat S, Yap TA. Advances in the development of molecularly targeted agents in non-small-cell lung cancer. Drugs. 2017;77:813–27.
Lategahn J, Keul M, Rauh D. Lessons to be learned: the molecular basis of kinase-targeted therapies and drug resistance in non-small cell lung cancer. Angew Chem Int Ed. 2018;57:2307–13.
Panda M, Biswal BK. Cell signaling and cancer: a mechanistic insight into drug resistance. Mol Biol Rep. 2019;46:5645–59.
Li BT, Drilon A, Johnson ML, Hsu M, Sima CS, McGinn C, et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol. 2016;27:154–9.
Zhang L, Wu H, Zhao M, Chang C, Lu Q. Clinical significance of miRNAs in autoimmunity. J Autoimmun. 2020;109:102438.
Arab A, Karimipoor M, Irani S, Kiani A, Zeinali S, Tafsiri E, et al. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet. 2017;216-217:150–8.
Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs -an update. Nat Rev Clin Oncol. 2018;15:541–63.
Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radio resistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45.
Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–82.
Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49:604–15.
Xu S, Chang Y, Wu Z, Li Y, Yuan R, Chai Y. One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens Bioelectron. 2020;149:111848.
Azzouzi S, Fredj Z, Turner APF, Ali MB, Mak WC. Generic neutravidin biosensor for simultaneous multiplex detection of microRNAs via electrochemically encoded responsive nanolabels. ACS Sensors. 2019;4:326–34.
Shen Z, He L, Wang W, Tan L, Gan N. Highly sensitive and simultaneous detection of microRNAs in serum using stir-bar assisted magnetic DNA nanospheres-encoded probes. Biosens Bioelectron. 2020;148:111831.
Tian R, Li Y, Bai J. Hierarchical assembled nanomaterial paper based analytical devices for simultaneously electrochemical detection of microRNAs. Anal Chim Acta. 2019;1058:89–96.
Wang J, Zheng C, Jiang Y, Zheng Z, Lin M, Lin Y, et al. One-step monitoring of multiple Enterovirus 71 infection-related MicroRNAs using Core−satellite structure of magnetic Nanobeadsand multicolor quantum dots. Anal Chem. 2020;92:830–7.
Xu S, Nie Y, Jiang L, Wang J, Xu G, Wang W, et al. Polydopamine nanosphere/gold nanocluster(AuNC)-based nanoplatform for dual color simultaneous detection of multiple tumor-related microRNAs with DNase-I-assisted target recycling amplification. Anal Chem. 2018;90:4039–45.
Cai X, Weng S, Guo R, Lin L, Chen W, Zheng Z, et al. Ratiometric electrochemical immunoassay based on internal reference value for reproducible and sensitive detection of tumor marker. Biosens Bioelectron. 2016;81:173–80.
Jin H, Gui R, Yu J, Lv W, Wang Z. Fabrication strategies, sensing modes and analytical applications of ratiometric electrochemical biosensors. Biosens Bioelectron. 2017;91:523–37.
Sun Y, Wang Y, Lau C, Chen G, Lu J. Hybridization-initiated exonuclease resistance strategy for simultaneous detection of multiple microRNAs. Talanta. 2018;190:248–54.
Pan L, Wang Z, Li Y, Xu F, Zhang Q, Zhang C. Nicking enzyme-controlled toehold regulation for DNA logic circuits. Nanoscale. 2017;9:18223–8.
Li Y, Chang Y, Yuan R, Chai Y. Highly efficient target recycling-based netlike Y-DNA for regulation of electrocatalysis toward methylene blue for sensitive DNA detection. ACS Appl Mater Inter. 2018;10:25213–8.
Yu H, Canoura J, Guntupalli B, Alkhamis O, Xiao Y. Sensitive detection of small-molecule targets using cooperative binding split aptamers and enzyme-sssisted target recycling. Anal Chem. 2018;90:1748–58.
Yuan C, Fang J, Duan Q, Yan Q, Guo J, Yuan T, et al. Two-layer three- dimensional DNA walker with highly integrated entropy-driven and enzyme-powered reactions for HIV detection. Biosens. Bioelectron. 2019;133:243–9.
Cui M, Xiao X, Zhao M, Zheng B. Detection of single nucleotide polymorphism by measuring extension kinetics with T7 exonuclease mediated isothermal amplification. Analyst. 2017;143:116–22.
Du YC, Cui YX, Li XY, Sun GY, Zhang YP, Tang AN, et al. Terminal deoxynucleotidyl transferase and T7 exonuclease-aided amplification strategy for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem. 2018;90:8629–34.
Xu J, Shi M, Huang H, Hu K, Chen W, Huang Y, et al. A fluorescent aptasensor based on single oligonucleotide-mediated isothermal quadratic amplification and graphene oxide fluorescence quenching for ultrasensitive protein detection. Analyst. 2018;143:3918–25.
Rosli NF, Fojtů M, Fisher AC, Pumera M. Graphene oxide nanoplatelets potentiate anticancer effect of cisplatin in human lung cancer cells. Langmuir. 2019;35:3176–82.
Weaver CL, LaRosa JM, Luo X, Cui XT. Electrically controlled drug delivery from graphene oxide nanocomposite films. ACS Nano. 2014;8:1834–43.
Oh HJ, Kim J, Park H, Chung S, Hwang DW, Lee DS. Graphene-oxide quenching-based molecular beacon imaging of exosome-mediated transfer of neurogenic miR-193a on microfluidic platform. Biosens Bioelectron. 2019;126:647–56.
Zhu QY, Zhang FR, Du Y, Zhang XX, Lu JY, Yao QF, et al. Graphene-based steganographicly aptasensing system for information computing, encryption and hiding, fluorescence sensing and in vivo imaging of fish pathogens. ACS Appl. Mater. Inter. 2019;11:8904–14.
Cui L, Lin X, Lin N, Song YL, Zhu Z, Chen X, et al. Graphene oxide-protected DNA probes for multiplex microRNA analysis in complex biological samples based on a cyclic enzymatic amplification method. Chem Commun. 2012;48:194–6.
Yong Jian Jiang YJ, Na Wang N, Feng Cheng F, Hua Rong Lin RH, Shu Jun Zhen SJ, Yuan Fang Li YF, et al. Dual energy transfer-based DNA / graphene oxide nanocomplex probe for highly robust and accurate monitoring of apoptosis-related microRNAs. Anal. Chem. 2020;92:11565–72.
Xiao X, Li YF, Huang CZ, Zhen SJ. A novel graphene oxide amplified fluorescence anisotropy assay with improved accuracy and sensitivity. Chem Commun. 2015;51:16080–3.
Zhen SJ, Xiao X, Li CH, Huang CZ. An enzyme-free DNA circuit-assisted graphene oxide enhanced fluorescence anisotropy assay for MicroRNA detection with improved sensitivity and selectivity. Anal Chem. 2017;89:8766–71.
Xiao JC, Hao XL, Miao CF, Li FL, Huang JY, Lin XH, et al. Determination of chondroitin sulfate in synovial fluid and drug by ratiometric fluorescence strategy based on carbon dots quenched FAM labeled ssDNA. Colloids Surf B. 2020;192:111030.
Cui L, Zhu Z, Lin N, Zhang H, Guan ZC, Yang CJY. A T7 exonuclease-assisted cyclic enzymatic amplification method coupled with rolling circle amplification: a dual-amplification strategy for sensitive and selective microRNA detection. Chem Commun. 2014;50:1576–8.
Zhang D, Lu M, Wang H. Fluorescence anisotropy analysis for mapping aptamer-protein interaction at the single nucleotide level. J Am Chem Soc. 2011;133:9188–91.
Li FL, Cai QQ, Hao XL, Zhao CF, Huang ZJ, Zheng YJ, et al. Insight into the DNA adsorption on nitrogen-doped positive carbon dots. RSC Adv. 2019;9:12462–9.
Huang JY, Li FL, Guo RB, Chen YY, Wang ZZ, Zhao CF, et al. A signal-on ratiometric fluorometric heparin assay based on the direct interaction between amino-modified carbon dots and DNA. Microchim Acta. 2018;185:260.
Zhang F, Wang S, Liu JW. Gold nanoparticles adsorb DNA and aptamer probes too strongly and a comparison with graphene oxide for biosensing. Anal Chem. 2019;91:14743–50.
Yuan YH, Wu YD, Chi BZ, Wen SH, Liang RP, Qiu JD. Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction. Biosens Bioelectron. 2017;97:325–31.
Han Y, Zhang F, Gong H, Cai C. Multifunctional G-quadruplex-based fluorescence probe coupled with DNA-templated AgNCs for simultaneous detection of multiple DNAs and MicroRNAs. Anal Chim Acta. 2019;1053:105–13.
Pang X, Qi J, Zhang Y, Ren Y, Su M, Jia B, et al. Ultrasensitive photoelectrochemical aptasensing of miR-155 using efficient and stable CH3NH3PbI3 quantum dots sensitized ZnO nanosheets as light harvester. Biosens Bioelectron. 2016;85:142–50.
Liang Z, Ou D, Sun D, Tong Y, Luo H, Chen Z. Ultrasensitive biosensor for microRNA-155 using synergistically catalytic nanoprobe coupled with improved cascade strand displacement reaction. Biosens Bioelectron. 2019;146:111744.
Acknowledgements
The authors gratefully acknowledge the financial support of the National Natural Science Foundation of China (21705021, 21775023 and 21904019), Joint Funds for the Innovation of Science and Technology, Fujian province (2017Y9121, 2019Y9011), Open project funded by Key laboratory of biological genetic resources from Ministry of Natural Resources (HY201703) and Medical Innovation Project of Fujian Province of China (2016-CX-44).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical standards and informed consent
This study conformed to the ethical guidelines of the Declaration of Helsinki and was approved by the Ethics Committee for Human Research, The First Affiliated Hospital of Fujian Medical University. Human serum samples used in this study do not have any identifying information about any of the participants, and all participants provided written informed consent.
Conflict of interest
The authors declare that no competing interests exist.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 625 kb)
Rights and permissions
About this article
Cite this article
Zheng, Y., Chen, J., Li, Y. et al. Dual-probe fluorescent biosensor based on T7 exonuclease-assisted target recycling amplification for simultaneous sensitive detection of microRNA-21 and microRNA-155. Anal Bioanal Chem 413, 1605–1614 (2021). https://doi.org/10.1007/s00216-020-03121-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-03121-6