Abstract
Resurrection plant Ramonda serbica is a suitable model to investigate vegetative desiccation tolerance. However, the detailed study of these mechanisms at the protein level is hampered by the severe tissue water loss, high amount of phenolics and polysaccharide, and possible protein modifications and aggregations during the extraction and purification steps. When applied to R. serbica leaves, widely used protein extraction protocols containing polyvinylpolypyrrolidone and ascorbate, as well as the phenol/SDS/buffer–based protocol recommended for recalcitrant plant tissues failed to eliminate persistent contamination and ensure high protein quality. Here we compared three protein extraction approaches aiming to establish the optimal one for both hydrated and desiccated R. serbica leaves. To evaluate the efficacy of these protocols by shotgun proteomics, we also created the first R. serbica annotated transcriptome database, available at http://www.biomed.unipd.it/filearrigoni/Trinity_Sample_RT2.fasta. The detergent-free phenol-based extraction combined with dodecyl-β-d-maltoside-assisted extraction enabled high-yield and high-purity protein extracts. The phenol-based protocol improved the protein-band resolution, band number, and intensity upon electrophoresis, and increased the protein yield and the number of identified peptides and protein groups by LC-MS/MS. Additionally, dodecyl-β-d-maltoside enabled solubilisation and identification of more membrane-associated proteins. The presented study paves the way for investigating the desiccation tolerance in R. serbica, and we recommend this protocol for similar recalcitrant plant material.
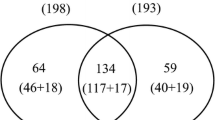
Graphical abstract




Similar content being viewed by others
Data availability
R. serbica transcriptome database, publically available at http://www.biomed.unipd.it/filearrigoni/Trinity_Sample_RT2.fasta
R. serbica protein fasta file: http://www.biomed.unipd.it/filearrigoni/Ramonda_Serbica_Protein_fasta_file.fasta
Abbreviations
- βME:
-
2-Mercaptoethanol
- CHAPS:
-
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulphonate
- DDM:
-
Dodecyl-β-d-maltoside
- DL:
-
Desiccated leaves
- EDTA:
-
Ethylenediaminetetraacetic acid
- FASP:
-
Filter-aided sample preparation
- HL:
-
Fully hydrated leaves
- LC:
-
Liquid chromatography
- MS:
-
Mass spectrometry
- PAGE:
-
Polyacrylamide gel electrophoresis
- PETC:
-
Photosynthetic electron transport chain
- PMSF:
-
Phenylmethylsulphonyl fluoride
- PS:
-
Photosystem
- PSM:
-
Peptide-spectrum match
- PVPP:
-
Water-insoluble polyvinylpolypyrrolidone
- RWC:
-
Relative water content
- SDS:
-
Sodium dodecyl sulphate
- TCA:
-
Trichloroacetic acid
References
Saravanan RS, Rose JK. A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics. 2004;4:2522–32.
Veljović-Jovanović S, Kukavica B, Stevanović B, Navari-Izzo F. Senescence-and drought-related changes in peroxidase and superoxide dismutase isoforms in leaves of Ramonda serbica. J Exp Bot. 2006;57:1759–68.
Veljović-Jovanović S, Kukavica B, Navari-Izzo F. Characterization of polyphenol oxidase changes induced by desiccation of Ramonda serbica leaves. Physiol Plant. 2008;132:407–16.
Farrant JM, Moore JP. Programming desiccation-tolerance: from plants to seeds to resurrection plants. Curr Opin Plant Biol. 2011;14:340–5.
Sgherri C, Stevanovic B, Navari-Izzo F. Role of phenolics in the antioxidative status of the resurrection plant Ramonda serbica during dehydration and rehydration. Physiol Plant. 2004;122:478–85.
Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;47:377–403.
Vidović M, Morina F, Veljović-Jovanović S. Stimulation of various phenolics in plants under ambient UV-B radiation. In: Singh VP, Singh S, Prasad SM, Parihar P, editors. UV-B radiation: from environmental stressor to regulator of plant growth. Chichester: Wiley-Blackwell; 2017. p. 9–56.
Sakakibara H, Honda Y, Nakagawa S, Ashida H, Kanazawa K. Simultaneous determination of all polyphenols in vegetables, fruits, and teas. J Agric Food Chem. 2003;51:571–81.
Kallinich C, Schefer S, Rohn S. Analysis of protein-phenolic compound modifications using electrochemistry coupled to mass spectrometry. Molecules. 2018;23:264.
Pierpoint WS. The extraction of enzymes from plant tissues rich in phenolic compounds. In: Cutler P, editor. Protein purification protocols. Methods in molecular biology, vol. 244: Humana Press; 2004. p. 65–74.
Vidović M, Ćuković K. Isolation of high-quality RNA from recalcitrant leaves of variegated and resurrection plants. 3 Biotech. 2020;10:286.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Chatterjee M, Gupta S, Bhar A, Das S. Optimization of an efficient protein extraction protocol compatible with two-dimensional electrophoresis and mass spectrometry from recalcitrant phenolic rich roots of chickpea (Cicer arietinum L.). Int. J Proteome. 2012;2012:536963.
Vidović M, Morina F, Milić-Komić S, Vuleta A, Zechmann B, Prokić L, et al. Characterisation of antioxidants in photosynthetic and non-photosynthetic leaf tissues of variegated Pelargonium zonale plants. Plant Biol. 2016;18:669–80.
Wang W, Vignani R, Scali M, Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27:2782–6.
Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–9.
Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Fryer HJ, Davis GE, Manthorpe M, Varon S. Lowry protein assay using an automatic microtiter plate spectrophotometer. Anal Biochem. 1986;153:262–6.
Brown RE, Jarvis KL, Hyland KJ. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem. 1989;180:136–9.
Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359.
Borgo C, Franchin C, Scalco S, Bosello-Travain V, Donella-Deana A, Arrigoni G, et al. Generation and quantitative proteomics analysis of CK2α/α’(−/−) cells. Sci Rep. 2017;7:42409.
Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, et al. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis. 2003;24:2369–75.
Lazarevic M, Siljak-Yakovlev S, Lazarevic P, Stevanovic B, Stevanovic V. Pollen and seed morphology of resurrection plants from the genus Ramonda (Gesneriaceae): relationship with ploidy level and relevance to their ecology and identification. Turk J Bot. 2013;37:872–85.
Liu J, Moyankova D, Lin CT, Mladenov P, Sun RZ, Djilianov D, et al. Transcriptome reprogramming during severe dehydration contributes to physiological and metabolic changes in the resurrection plant Haberlea rhodopensis. BMC Plant Biol. 2018;18:351.
Zhu Y, Wang B, Phillips J, Zhang ZN, Du H, Xu T, et al. Global transcriptome analysis reveals acclimation-primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiol. 2015;56:1429–41.
Rodriguez MCS, Edsgärd D, Hussain SS, Alquezar D, Rasmussen M, Gilbert T, et al. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 2010;63:212–28.
Xiao L, Yang G, Zhang L, Yang X, Zhao S, Ji Z, et al. The resurrection genome of Boea hygrometrica: a blueprint for survival of dehydration. Proc Natl Acad Sci U S A. 2015;112:5833–7.
Wu X, Gong F, Wang W. Protein extraction from plant tissues for 2 DE and its application in proteomic analysis. Proteomics. 2014;14:645–58.
Raak N, Abbate RA, Lederer A, Rohm H, Jaros D. Size separation techniques for the characterisation of cross-linked casein: a review of methods and their applications. Separations. 2018;5:14.
Vogt EI, Kupfer VM, Vogel RF, Niessen L. A novel preparation technique of red (sparkling) wine for protein analysis. EuPA Open Proteom. 2016;11:16–9.
Rezadoost MH, Kordrostami M, Kumleh HH. An efficient protocol for isolation of inhibitor-free nucleic acids even from recalcitrant plants. 3 Biotech. 2016;6:61.
Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B. Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics. 2005;5:2497–507.
Gray JC. Absorption of polyphenols by polyvinylpyrrolidone and polystyrene resins. Phytochemistry. 1978;17:495–7.
Jin X, Zhu L, Tao C, Xie Q, Xu X, Chang L, et al. An improved protein extraction method applied to cotton leaves is compatible with 2-DE and LC-MS. BMC Genomics. 2019;20:285.
Sakihama Y, Cohen MF, Grace SC, Yamasaki H. Plant phenolic antioxidant and prooxidant activities: phenolics-induced oxidative damage mediated by metals in plants. Toxicology. 2002;177:67–80.
Davies MJ. Protein oxidation and peroxidation. Biochem J. 2016;473:805–25.
Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201.
Röhrig H, Colby T, Schmidt J, Harzen A, Facchinelli F, Bartels D. Analysis of desiccation-induced candidate phosphoproteins from Craterostigma plantagineum isolated with a modified metal oxide affinity chromatography procedure. Proteomics. 2008;8:3548–60.
Wu X, Xiong E, Wang W, Scali M, Cresti M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat Protoc. 2014;9:362–74.
Singh N, Jain N, Kumar R, Jain A, Singh NK, Rai V. A comparative method for protein extraction and 2-D gel electrophoresis from different tissues of Cajanus cajan. Front Plant Sci. 2015;6:606.
Jiang G, Wang Z, Shang H, Yang W, Hu Z, Phillips J, et al. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta. 2007;225:1405.
Wang X, Chen S, Zhang H, Shi L, Cao F, Guo L, et al. Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J Proteome Res. 2010;9:6561–77.
Ingle RA, Schmidt UG, Farrant JM, Thomson JA, Mundree SG. Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant Cell Environ. 2007;30:435–46.
Lee J, Koh HJ. A label-free quantitative shotgun proteomics analysis of rice grain development. Proteome Sci. 2011;9:61.
Feroz H, Kwon H, Peng J, Oh H, Ferlez B, Baker CS, et al. Improving extraction and post-purification concentration of membrane proteins. Analyst. 2018;143:1378–86.
Stetsenko A, Guskov A. An overview of the top ten detergents used for membrane protein crystallization. Crystals. 2017;7:197.
Acknowledgements
The authors would like to acknowledge the technical help provided Dr. Anna Rita Trentin (DAFNAE, University of Padova) and Ilaria Battisti (Proteomics Center University of Padova and Azienda Ospedaliera di Padova). The authors wish to thank the Cassa di Risparmio di Padova e Rovigo (Cariparo) Holding for funding the acquisition of the LTQ-Orbitrap XL mass spectrometer.
Funding
This work was funded by the Ministry of Education, Science and Technological Development, Republic of Serbia (contract no. 451-03-68/2020-14/200053), the Science Fund of the Republic of Serbia (PROMIS project LEAPSyn-SCI, grant number 6039663), and the University of Padova (grant number BIRD189887/18 to G.A). M.Vidović received financial support from COST Action BM1405 (STSM-BM1405-190218-092344 and STSM-BM1405-190317-080965).
Author information
Authors and Affiliations
Contributions
M. Vidović and G. Arrigoni designed the research. F. Morina, S. Veljović-Jovanović, and M.Vidović collected the material and conducted the experiment. F. Morina extracted and purified total RNA. M. Vidović tested the protein extraction procedures. M. Vidović, C. Franchin, and G. Arrigoni performed peptide digestion, purification, and proteomic analysis. M. Vidović and G. Arrigoni analysed the data, and together with F. Morina, A. Masi, and S. Veljović-Jovanović discussed the results and wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vidović, M., Franchin, C., Morina, F. et al. Efficient protein extraction for shotgun proteomics from hydrated and desiccated leaves of resurrection Ramonda serbica plants. Anal Bioanal Chem 412, 8299–8312 (2020). https://doi.org/10.1007/s00216-020-02965-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02965-2