Abstract
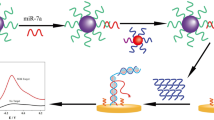
This paper reports a simple electrochemical strategy for the determination of microRNAs (miRNAs) using a commercial His-Tag-Zinc finger protein (His-Tag-ZFP) that binds preferably (but non-sequence specifically) RNA hybrids over ssRNAs, ssDNAs, and dsDNAs. The strategy involves the use of magnetic beads (His-Tag-Isolation-MBs) as solid support to capture the conjugate formed in homogenous solution between His-Tag-ZFP and the dsRNA homohybrid formed between the target miRNA (miR-21 selected as a model) and a biotinylated synthetic complementary RNA detector probe (b-RNA-Dp) further conjugated with a streptavidin–horseradish peroxidase (Strep–HRP) conjugate. The electrochemical detection is carried out by amperometry at disposable screen-printed carbon electrodes (SPCEs) (− 0.20 V vs Ag pseudo-reference electrode) upon magnetic capture of the resultant magnetic bioconjugates and H2O2 addition in the presence of hydroquinone (HQ). The as-prepared biosensor exhibits a dynamic concentration range from 3.0 to 100 nM and a detection limit (LOD) of 0.91 nM for miR-21 in just ~ 2 h. An acceptable discrimination was achieved between the target miRNA and other non-target nucleic acids (ssDNA, dsDNA, ssRNA, DNA–RNA, miR-122, miR-205, and single central- or terminal-base mismatched sequences). The biosensor was applied to the analysis of miR-21 from total RNA (RNAt) extracted from epithelial non-tumorigenic and adenocarcinoma breast cells without target amplification, pre-concentration, or reverse transcription steps. The versatility of the methodology due to the ZFP’s non-sequence-specific binding behavior makes it easily extendable to determine any target RNA only by modifying the biotinylated detector probe.





Similar content being viewed by others
References
Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25:137–47.
Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer. Int J Oncol. 2012;41:1897–912.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70.
Peng Y, Croce CM. The role of microRNAs in human cancer. Signal transduction and targeted therapy. 2016;1:15004. https://doi.org/10.1038/sigtrans.2015.4.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9.
Masud MK, Umer M, Hossain SA, Yamauchi Y, Nguyen NT, Shiddiky MJA. Nanoarchitecture frameworks for electrochemical miRNA detection. Trends in Biomedical Sciences. 2019;44:433–52.
Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos JZ. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1:155–61.
Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA. 2006;12:913–20.
Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4.
Li J, Yao B, Huang H, Wang Z, Sun C, Fan Y, et al. Real-time polymerase chain reaction microRNA detection based on enzymatic stem-loop probes ligation. Anal Chem. 2009;81:5446–51.
Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50:298–301.
Hunt EA, Broyles D, Head T, Deo SK. MicroRNA detection: current technology and research strategies. Annu Rev Anal Chem. 2015;8:217–37.
Chen YX, Huang KJ, Niu KX. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens Bioelectron. 2018;99:612–24.
Islam MN, Masud MK, Nguyen NT, Gopalan V, Alamri HR, Alothman ZA, et al. Gold-loaded nanoporous ferric oxide nanocubes for electrocatalytic detection of microRNA at attomolar level. Biosens Bioelectron. 2018;101:275–81.
Voccia D, Palchetti I. Electrochemical biosensors for miRNA detection. In: Erdmann V, Jurga S, Barciszewski J, editors. RNA and DNA diagnostics. Cham.: RNA technologies. Springer; 2015. p. 1–19.
Campuzano S, Pedrero M, Pingarron JM. Viral protein-based bioanalytical tools for small RNA biosensing. Trends Anal Chem. 2016;79:335–43.
Planell-Saguer MD, Rodicio MC. Analytical aspects of microRNA in diagnostics: a review. Anal Chim Acta. 2011;699:134–52.
Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. MicroRNA: function, detection, and bioanalysis. Chem Rev. 2013;113:6207–33.
Wu J, Campuzano S, Halford C, Haake DA, Wang J. Ternary surface monolayers for ultrasensitive (zeptomole) amperometric detection of nucleic acid hybridization without signal amplification. Anal Chem. 2010;82:8830–7.
Campuzano S, Kuralay F, Lobo-Castañón MJ, Bartošik M, Vyavahare K, Paleček E, et al. Ternary monolayers as DNA recognition interfaces for direct and sensitive electrochemical detection in untreated clinical samples. Biosens Bioelectron. 2011;26:3577–83.
Wark W, Lee HJ, Corn RM. Multiplexed detection methods for profiling microRNA expression in biological samples. Angew Chem Int Ed. 2008;47:644–52.
Lee JM, Jung Y. Two-temperature hybridization for microarray detection of label-free microRNAs with attomole detection and superior specificity. Angew Chem Int Ed. 2011;50:12487–90.
Campuzano S, Torrente-Rodríguez RM, López-Hernández E, Conzuelo F, Granados R, Sanchez-Puelles JM, et al. Magnetobiosensors based on viral protein p19 for microRNA determination in cancer cells and tissues. Angew Chem Int Ed. 2014;53:6168–71.
Torrente-Rodríguez RM, Ruiz-Valdepeñas Montiel V, Campuzano S, Farchado-Dinia M, Barderas R, San Segundo-Acosta P, et al. Fast electrochemical miRNAs determination in cancer cells and tumor tissues with antibody-functionalized magnetic microcarriers. ACS Sens. 2016;1:896–903.
Vargas E, Torrente-Rodríguez RM, Ruiz-Valdepeñas Montiel V, Povedano E, Pedrero M, Montoya JJ, et al. Magnetic beads-based sensor with tailored sensitivity for rapid and single-step amperometric determination of miRNAs. Int J Mol Sci. 2017;18:2151. https://doi.org/10.3390/ijms18112151.
Fang CS, Kim KS, Yu B, Jo S, Kim MS, Yang H. Ultrasensitive electrochemical detection of miRNA-21 using a zinc finger protein specific to DNA–RNA hybrids. Anal Chem. 2017;89:2024–31.
Fang CS, Kim K, Ha DT, Kim M, Yang H. Washing-free electrochemical detection of amplified double-stranded DNAs using a zinc finger protein. Anal Chem. 2018;90:4776–82.
Ren Y, Deng H, Shen W, Gao ZA. Highly sensitive and selective electrochemical biosensor for direct detection of microRNAs in serum. Anal Chem. 2013;85:4784–9.
Campuzano S, Yánez-Sedeño P, Pingarrón JM. Electrochemical biosensing of microribonucleic acids using antibodies and viral proteins with affinity for ribonucleic acid duplexes. Electrochim Acta. 2017;230:271–8.
Camenisch TD, Brilliant MH, Segal DJ. Critical parameters for genome editing using zinc finger nucleases. Mini-Rev Med Chem. 2008;8:669–76.
Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604.
Kim MS, Stybayeva G, Lee JY, Revzin A, Segal DJ. A zinc finger protein array for the visual detection of specific DNA sequences for diagnostic applications. Nucleic Acids Res. 2011;39:e29. https://doi.org/10.1093/nar/gkq1214.
Hiraoka D, Yoshida W, Abe K, Wakeda H, Hata K, Ikebukuro K. Development of a method to measure DNA methylation levels by using methyl CpG-binding protein and luciferase-fused zinc finger protein. Anal Chem. 2012;84:8259–64.
Takano E, Shimura N, Akiba T, Kitayama Y, Sunayama H, Abe K, et al. Pipette tip biosensors for bacterial double-stranded DNA using bioluminescence induced by zinc finger luciferase. Microchim Acta. 2017;184:1595–601.
Lee J, Tatsumi A, Abe K, Yoshida W, Sodea K, Ikebukuro K. Electrochemical detection of pathogenic bacteria by using a glucose dehydrogenase fused zinc finger protein. Analyst. 2014;6:4991–4.
Lee J, Yoshida W, Abe K, Nakabayashi K, Wakeda H, Hata K, et al. Development of an electrochemical detection system for measuring DNA methylation levels using methyl CpG-binding protein and glucose dehydrogenase-fused zinc finger protein. Biosens Bioelectron. 2017;93:118–23.
Kim MS, Kim J. Multiplexed detection of pathogen-specific DNA using engineered zinc finger proteins without target amplification. Anal Methods. 2016;8:6696–700.
Noh S, Ha DT, Yang H, Kim MS. Sensitive and direct electrochemical detection of double-stranded DNA utilizing alkaline phosphatase-labelled zinc finger proteins. Analyst. 2015;140:3947–52.
Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–35.
Zouari M, Campuzano S, Pingarrón JM, Raouafi N. Competitive RNA-RNA hybridization-based integrated nanostructured disposable electrode for highly sensitive determination of miRNAs in cancer cells. Biosens. Bioelectron. 2017;91:40–5.
Vargas E, Povedano E, Ruiz-Valdepeñas Montiel V, Torrente-Rodríguez RM, Zouari M, Montoya JJ, et al. Single-step incubation determination of miRNAs in cancer cells using an amperometric biosensor based on competitive hybridization onto magnetic beads. Sensors. 2018;18:863. https://doi.org/10.3390/s18030863.
Kilic T, Topkaya SN, Ariksoysal DO, Ozsoz M, Ballar P, Erac Y, et al. Electrochemical based detection of microRNA, mir21 in breast cancer cells. Biosens Bioelectron. 2012;38:195–201.
Eguílaz M, Moreno-Guzmán M, Campuzano S, González-Cortés A, Yáñez-Sedeño P, Pingarrón JM. An electrochemical immunosensor for testosterone using functionalized magnetic beads and screen-printed carbon electrodes. Biosens Bioelectron. 2010;26:517–22.
Conzuelo F, Gamella M, Campuzano S, Reviejo AJ, Pingarrón JM. Disposable amperometric magneto-immunosensor for direct detection of tetracyclines antibiotics residues in milk. Anal Chim Acta. 2012;737:29–36.
Gamella M, Campuzano S, Conzuelo F, Reviejo AJ, Pingarrón JM. Amperometric magnetoimmunosensors for direct determination of D-dimer in human serum. Electroanalysis. 2012;24:2235–43.
Moyo M. Horseradish peroxidase biosensor to detect zinc ions in aqueous solutions. Open J Appl Biosens. 2014;3:1–7.
Li F, Peng J, Wang J, Tang H, Tan L, Xie Q, et al. Carbon nanotube-based label-free electrochemical biosensor for sensitive detection of miRNA-24. Biosens Bioelectron. 2014;54:158–64.
Jin J, Cid M, Poole CB, McReynolds LA. Protein mediated miRNA detection and siRNA enrichment using p19. BioTechniques. 2010;48:xvii–xxiii.
Gillespie P, Ladame S, O’Hare D. Molecular methods in electrochemical microRNA detection. Analyst. 2019;144:114–29.
Li L, Feng J, Liu H, Li Q, Tong L, Tang B. Two-color imaging of microRNA with enzyme-free signal amplification via hybridization chain reactions in living cells. Chem Sci. 2016;7:1940–5.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60.
Funding
The financial support of the CTQ2015-64402-C2-1-R (Spanish Ministerio de Economía y Competitividad) and RTI2018-096135-B-I00 (Ministerio de Ciencia, Innovación y Universidades) Research Projects and the TRANSNANOAVANSENS-CM Program from the Comunidad de Madrid (Grant S2018/NMT-4349) are gratefully acknowledged, and predoctoral contracts from the Spanish Ministerio de Economía y Competitividad (EP) and Universidad Complutense de Madrid (VRVM) are also gratefully acknowledged. VS was financially supported by a postdoctoral contract type Art 83. LOU with MiRNAx Biosens. S.L. company. MB and LJ would like to acknowledge financial support from the projects of Czech Science Foundation 17-08971S, MEYS – NPS I – LO1413, and MH CZ - DRO (MMCI, 00209805).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Published in the topical collection Euroanalysis XX with guest editor Sibel A. Ozkan.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 583 KB)
Rights and permissions
About this article
Cite this article
Povedano, E., Ruiz-Valdepeñas Montiel, V., Gamella, M. et al. A novel zinc finger protein–based amperometric biosensor for miRNA determination. Anal Bioanal Chem 412, 5031–5041 (2020). https://doi.org/10.1007/s00216-019-02219-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-02219-w