Abstract
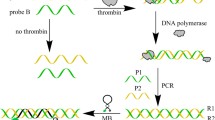
This work proposed a homogenous fluorescence assay for proteins, based on the target-triggered proximity DNA hybridization in combination with strand displacement amplification (SDA). It benefited from target-triggered proximity DNA hybridization to specifically recognize the target and SDA making recycling signal amplification. The system included a molecular beacon (MB), an extended probe (EP), and an assistant probe (AP), which were not self-assembly in the absence of target proteins, due to the short length of the designed complementary sequence among MB, EP, and AP. Upon addition of the target proteins, EP and AP are bound to the target proteins, which induced the occurrence of proximity hybridization between MB, EP, and AP and followed by strand displacement amplification. Through the primer extension, a tripartite complex of probes and target was displaced and recycled to hybridize with another MB, and the more opened MB enabled the detection signal to amplify. Under optimum conditions, it was used for the detection of streptavidin and thrombin. Fluorescence intensity was proportional to the concentration of streptavidin and thrombin in the range of 0.2–30 and 0.2–35 nmol/L, respectively. Furthermore, this fluorescent method has a good selectivity, in which the fluorescence intensity of thrombin was ~37-fold or even larger than that of the other proteins at the same concentration. It is a new and simple method for SDA-involved target protein detection and possesses a great potential for other protein detection in the future.

A homogenous assay for protein detection is based on proximity DNA hybridization and strand displacement amplification reaction.




Similar content being viewed by others
References
Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–9. doi:10.1038/nature06916.
Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462(7272):461–4. doi:10.1038/nature08605.
Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). ClinChem. 2005;51(12):2415–8. doi:10.1373/clinchem.2005.051532.
Rusling JF. Multiplexed electrochemical protein detection and translation to personalized cancer diagnostics. Anal Chem. 2013;85(11):5304–10. doi:10.1021/ac401058v.
Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20(5):473–7. doi:10.1038/nbt0502-473.
Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi:10.1038/nmeth947.
Schallmeiner E, Oksanen E, Ericsson O, Spangberg L, Eriksson S, Stenman UH, Pettersson K, Landegren U. Sensitive protein detection via triple-binder proximity ligation assays. Nat Methods. 2007;4(2):135–7. doi:10.1038/nmeth974.
Li F, Zhang H, Wang Z, Li X, Li X, Le XC. Dynamic DNA assemblies mediated by binding-induced DNA strand displacement. J Am ChemSoc. 2013;135(7):2443–6. doi:10.1021/ja311990w.
Li F, Lin Y, Le XC. Binding-induced formation of DNA three-way junctions and its application to protein detection and DNA strand displacement. Anal Chem. 2013;85(22):10835–41. doi:10.1021/ac402179a.
Wu H, Zhang K, Liu Y, Wang H, Wu J, Zhu F, Zou P. Binding-induced and label-free colorimetric method for protein detection based on autonomous assembly of hemin/G-quadruplexDNAzyme amplification strategy. Biosens Bioelectron. 2015;64:572–8. doi:10.1016/j.bios.2014.09.096.
Zhang L, Zhang K, Liu G, Liu M, Liu Y, Li J. Label-free nanopore proximity bioassay for platelet-derived growth factor detection. Anal Chem. 2015;87(11):5677–82. doi:10.1021/acs.analchem.5b00791.
Zong C, Wu J, Liu M, Yang L, Liu L, Yan F, Ju H. Proximity hybridization-triggered signal switch for homogeneous chemiluminescent bioanalysis. Anal Chem. 2014;86(11):5573–8. doi:10.1021/ac501091n.
Wei Y, Zhou W, Liu J, Chai Y, Xiang Y, Yuan R. Label-free and homogeneous aptamer proximity binding assay for fluorescent detection of protein biomarkers in human serum. Talanta. 2015;141:230–4. doi:10.1016/j.talanta.2015.04.005.
Ren K, Wu J, Ju H, Yan F. Target-driven triple-binder assembly of MNAzyme for amplified electrochemical immunosensing of protein biomarker. Anal Chem. 2015;87(3):1694–700. doi:10.1021/ac504277z.
Liu B, Chen J, Wei Q, Zhang B, Zhang L, Tang D. Target-regulated proximity hybridization with three-way DNA junction for in situ enhanced electronic detection of marine biotoxin based on isothermal cycling signal amplification strategy. Biosens Bioelectron. 2015;69:241–8. doi:10.1016/j.bios.2015.02.040.
Zhang H, Li F, Dever B, Wang C, Li XF, Le XC. Assembling DNA through affinity binding to achieve ultrasensitive protein detection. Angew Chem Int Ed Engl. 2013;52(41):10698–705. doi:10.1002/anie.201210022.
Li X, Huang Y, Guan Y, Zhao M, Li Y. Universal molecular beacon-based tracer system for real-time polymerase chain reaction. Anal Chem. 2006;78(22):7886–90. doi:10.1021/ac061518+.
Yang L, Fung CW, Cho EJ, Ellington AD. Real-time rolling circle amplification for protein detection. Anal Chem. 2007;79(9):3320–9. doi:10.1021/ac062186b.
Liu X, Freeman R, Willner I. Amplified fluorescence aptamer-based sensors using exonuclease III for the regeneration of the analyte. Chem-Eur J. 2012;18(8):2207–11. doi:10.1002/chem.201103342.
Xue L, Zhou X, Xing D. Sensitive and homogeneous protein detection based on target-triggered aptamer hairpin switch and nicking enzyme assisted fluorescence signal amplification. Anal Chem. 2012;84(8):3507–13. doi:10.1021/ac2026783.
Luo M, Li N, Liu Y, Chen C, Xiang X, Ji X, He Z. Highly sensitive and multiple DNA biosensor based on isothermal strand-displacement polymerase reaction and functionalized magnetic microparticles. Biosens Bioelectron. 2014;55:318–23. doi:10.1016/j.bios.2013.11.066.
Wang F, Freage L, Orbach R, Willner I. Autonomous replication of nucleic acids by polymerization/nicking enzyme/DNAzyme cascades for the amplified detection of DNA and the aptamer–cocaine complex. Anal Chem. 2013;85(17):8196–203. doi:10.1021/ac4013094.
Tan E, Wong J, Nguyen D, Zhang Y, Erwin B, Van Ness LK, Baker SM, Galas DJ, Niemz A. Isothermal DNA amplification coupled with DNA nanosphere-based colorimetric detection. Anal Chem. 2005;77(24):7984–92. doi:10.1021/ac051364i.
Van Ness J, Van Ness LK, Galas DJ. Isothermal reactions for the amplification of oligonucleotides. Proc Natl AcadSci U S A. 2003;100(8):4504–9. doi:10.1073/pnas.0730811100.
Guo Q, Yang X, Wang K, Tan W, Li W, Tang H, Li H. Sensitive fluorescence detection of nucleic acids based on isothermal circular strand-displacement polymerization reaction. Nucleic Acids Res. 2009;37(3):e20. doi:10.1093/nar/gkn1024.
Duan R, Zuo X, Wang S, Quan X, Chen D, Chen Z, Jiang L, Fan C, Xia F. Quadratic isothermal amplification for the detection of microRNA. Nat Protoc. 2014;9(3):597–607. doi:10.1038/nprot.2014.036.
Huang Y, Liu X, Huang H, Qin J, Zhang L, Zhao S, Chen ZF, Liang H. Attomolar detection of proteins via cascade strand-displacement amplification and polystyrene nanoparticle enhancement in fluorescence polarization aptasensors. Anal Chem. 2015;87(16):8107–14. doi:10.1021/ac5041692.
Wang H, Wang Y, Liu S, Yu J, Xu W, Guo Y, Huang J. Target–aptamer binding triggered quadratic recycling amplification for highly specific and ultrasensitive detection of antibiotics at the attomole level. Chem Commun. 2015;51(39):8377–80. doi:10.1039/C5CC01473E.
Li Y, Miao X, Ling L. Triplex DNA: a new platform for polymerase chain reaction-based biosensor. Sci Rep. 2015;5:13010. doi:10.1038/srep13010.
Shao K, Wang B, Ye S, Zuo Y, Wu L, Li Q, Lu Z, Tan X, Han H. Signal-amplified near-infrared ratiometric electrochemiluminescence aptasensor based on multiple quenching and enhancement effect of graphene/gold nanorods/G-quadruplex. Anal Chem. 2016;88(16):8179–87. doi:10.1021/acs.analchem.6b01935.
Lin P, Chen R, Lee C, Chang Y, Chen C, Chen W. Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry. Colloids Surf B: Biointerfaces. 2011;88(2):552–8. doi:10.1016/j.colsurfb.2011.07.032.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21375153) and the Natural Science Foundation of Guangdong Province (Grant No. 2016A030310188).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
The study was approved by the Ethics Committee of Sun Yat-sen University, and informed consent was obtained from all individuals providing blood serum samples.
Electronic supplementary material
ESM 1
(PDF 290 kb)
Rights and permissions
About this article
Cite this article
Zhang, M., Li, R. & Ling, L. Homogenous assay for protein detection based on proximity DNA hybridization and isothermal circular strand displacement amplification reaction. Anal Bioanal Chem 409, 4079–4085 (2017). https://doi.org/10.1007/s00216-017-0356-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0356-0