Abstract
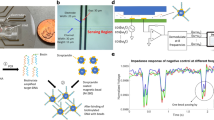
This research demonstrates an integrated microfluidic titration assay to characterize the cation concentrations in working buffer to rapidly optimize the signal-to-noise ratio (SNR) of molecular beacons (MBs). The “Microfluidic Droplet Array Titration Assay" (MiDATA) integrated the functions of sample dilution, sample loading, sample mixing, fluorescence analysis, and re-confirmation functions all together in a one-step process. It allows experimentalists to arbitrarily change sample concentration and acquire SNR measurements instantaneously. MiDATA greatly reduces sample dilution time, number of samples needed, sample consumption, and the total titration time. The maximum SNR of molecular beacons is achieved by optimizing the concentrations of the monovalent and divalent cation (i.e., Mg2+ and K+) of the working buffer. MiDATA platform is able to reduce the total consumed reagents to less than 50 μL, and decrease the assay time to less than 30 min. The SNR of the designated MB is increased from 20 to 126 (i.e., enhanced the signal 630 %) using the optimal concentration of MgCl2 and KCl determined by MiDATA. This novel microfluidics-based titration method is not only useful for SNR optimization of molecular beacons but it also can be a general method for a wide range of fluorescence resonance energy transfer (FRET)-based molecular probes.

The concentration of monovalent (K+) and divalent (Mg2+) cation in working buffer influences the signal-to-noise ratio (SNR) of molecular beacon (MB). Thus, optimizing the cationic concentrations in working buffer is necessary to achieve optimal SNR of MB assays for sensitive nucleic acids analysis.





Similar content being viewed by others
References
Tyagi S, Bratu DP, Kramer FR (1998) Multicolor molecular beacons for allele discrimination. Nat Biotechnol 16(1):49–53
Marras SAE, Kramer FR, Tyagi S (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal Biomol Eng 14(5–6):151–156
Vet JAM, Majithia AR, Marras SAE, Tyagi S, Dube S, Poiesz BJ, Kramer FR (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci U S A 96(11):6394–6399
Kwok PY (2001) Methods for genotyping single nucleotide polymorphisms. Annu Rev Genom Hum Genet 2:235–258
Syvanen AC (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet 2(12):930–942
Fang XH, Li JJ, Tan WH (2000) Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal Chem 72(14):3280–3285
Tung CH, Mahmood U, Bredow S, Weissleder R (2000) In vivo imaging of proteolytic enzyme activity using a novel molecular reporter. Cancer Res 60(17):4953–4958
Chen W, Martinez G, Mulchandani A (2000) Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem 280(1):166–172
Vogelstein B, Kinzler KW (1999) Digital PCR. Proc Natl Acad Sci U S A 96(16):9236–9241
Sokol DL, Zhang XL, Lu PZ, Gewitz AM (1998) Real time detection of DNA RNA hybridization in living cells. Proc Natl Acad Sci U S A 95(20):11538–11543
Matsuo T (1998) In situ vizualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim Biophys Acta Gen Subj 1379(2):178–184
Fang XH, Mi YM, Li JWJ, Beck T, Schuster S, Tan WH (2002) Molecular beacons–fluorogenic probes for living cell study. Cell Biochem Biophys 37(2):71–81
Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei-Iida Y, Kondo S, Ishibashi K (2000) Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys J 78(6):3260–3274
Yao G, Fang XH, Yokota H, Yanagida T, Tan WH (2003) Monitoring molecular beacon DNA probe hybridization at the single-molecule level. Chem Eur J 9(22):5686–5692
Tyagi S, Kramer FR (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14(3):303–308
Stryer L, Haugland RP (1967) Energy transfer—a spectroscopic ruler. Proc Natl Acad Sci USA 58(2):719–726
Deniz AA, Dahan M, Grunwell JR, Ha TJ, Faulhaber AE, Chemla DS, Weiss S, Schultz PG (1999) Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc Natl Acad Sci U S A 96(7):3670–3675
Stryer L (1978) Fluorescence energy-transfer as a spectroscopic ruler. Ann Rev Biochem 47:819–846
Tan WH, Wang KM, Drake TJ (2004) Molecular beacons. Curr Opin Chem Biol 8(5):547–553
Hardin CC, Watson T, Corregan M, Bailey C (1992) Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of D(Cgcg3gcg). Biochemistry 31(3):833–841
Williamson JR (1994) G-quartet structures in telomeric DNA. Ann Rev Biophys Biomol Struc 23:703–730
Tsourkas A, Behlke MA, Bao G (2002) Structure–function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res 30(19):4208–4215
Tsourkas A, Behlke MA, Rose SD, Bao G (2003) Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res 31(4):1319–1330
Bonnet G, Krichevsky O, Libchaber A (1998) Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci U S A 95(15):8602–8606
Bonnet G, Tyagi S, Libchaber A, Kramer FR (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci U S A 96(11):6171–6176
Yao G, Tan WH (2004) Molecular beacon-based array for sensitive DNA analysis. Anal Biochem 331(2):216–223
Gao Y, Wolf LK, Georgiadis RM (2006) Secondary structure effects on DNA hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res 34(11):3370–3377
Du H, Strohsahl CM, Camera J, Miller BL, Krauss TD (2005) Sensitivity and specificity of metal surface-immobilized "molecular beacon" biosensors. J Am Chem Soc 127(21):7932–7940
Fang XH, Liu XJ, Schuster S, Tan WH (1999) Designing a novel molecular beacon for surface-immobilized DNA hybridization studies. J Am Chem Soc 121(12):2921–2922
Reynolds O (1883) An experimental investigation of the circumstances which determines whether the motion of water shall be direct or sinuous, and of the law of resistance in parallel channels. Phil Trans R Soc 174:935–982
Kenis PJA, Ismagilov RF, Whitesides GM (1999) Microfabrication inside capillaries using multiphase laminar flow patterning. Science 285(5424):83–85
Liu RH, Yang JN, Pindera MZ, Athavale M, Grodzinski P (2002) Bubble-induced acoustic micromixing. Lab Chip 2(3):151–157
Stroock AD, Dertinger SKW, Ajdari A, Mezic I, Stone HA, Whitesides GM (2002) Chaotic mixer for microchannels. Science 295(5555):647–651
Hsieh ATH, Hori N, Massoudi R, Pan PJH, Sasaki H, Lin YA, Lee AP (2009) Nonviral gene vector formation in monodispersed picolitre incubator for consistent gene delivery. Lab Chip 9(18):2638–2643
Tice JD, Song H, Lyon AD, Ismagilov RF (2003) Formation of droplets and mixing in multiphase microfluidics at low values of the Reynolds and the capillary numbers. Langmuir 19(22):9127–9133
Liau A, Karnik R, Majumdar A, Cate JHD (2005) Mixing crowded biological solutions in milliseconds. Anal Chem 77(23):7618–7625. doi:10.1021/Ac050827h
Hsieh ATH, Pan PJH, Lee AP (2009) Rapid label-free DNA analysis in picoliter microfluidic droplets using FRET probes. Microfluidics Nanofluidics 6(3):391–401
Song H, Ismagilov RF (2003) Millisecond kinetics on a microfluidic chip using nanoliters of reagents. J Am Chem Soc 125(47):14613–14619
Muradoglu M, Stone HA (2005) Mixing in a drop moving through a serpentine channel: a computational study. Phys Fluids 17(7):073305–073309
Gee KR, Brown KA, Chen WNU, Bishop-Stewart J, Gray D, Johnson I (2000) Chemical and physiological characterization of fluo-4 Ca2 + -indicator dyes. Cell Calcium 27(2):97–106
Shattuckeidens D, Mcclure M, Simard J, Labrie F, Narod S, Couch F, Hoskins K, Weber B, Castilla L, Erdos M, Brody L, Friedman L, Ostermeyer E, Szabo C, King MC, Jhanwar S, Offit K, Norton L, Gilewski T, Lubin M, Osborne M, Black D, Boyd M, Steel M, Ingles S, Haile R, Lindblom A, Olsson H, Borg A, Bishop DT, Solomon E, Radice P, Spatti G, Gayther S, Ponder B, Warren W, Stratton M, Liu QY, Fujimura F, Lewis C, Skolnick MH, Goldgar DE (1995) A collaborative survey of 80 mutations in the Brca1 breast cancer and ovarian cancer susceptibility gene— implications for presymptomatic testing and screening. JAMA 273(7):535–541
Duffy DC, McDonald JC, Schueller OJA, Whitesides GM (1998) Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Analy Chem 70(23):4974–4984
Acknowledgments
This work was supported in part by the Defense Advanced Research Projects Agency (DARPA) N/MEMS S&T Fundamentals program under grant number HR001-06-1-0500 issued to the Micro/Nano Fluidics Fundamentals Focus (MF3) Center; and The National Aeronautics and Space Administration (NASA) through the University Affiliated Research Center under grant number NAS2-03144.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 190 kb)
Rights and permissions
About this article
Cite this article
Hsieh, A.TH., Pan, P.J. & Lee, A.P. A real-time characterization method to rapidly optimize molecular beacon signal for sensitive nucleic acids analysis. Anal Bioanal Chem 406, 3059–3067 (2014). https://doi.org/10.1007/s00216-014-7721-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-7721-z