Abstract
In this work, we establish a methodology for comparing the efficiencies of different hydrazide labels for detecting protein carbonyls. We have chosen acrolein-modified human serum albumin as a model. This system provides a convenient means of reproducibly generating carbonylated protein. Five hydrazide-based labels were tested. Three carry a biotin affinity tag, and the others are simple fatty acid hydrazides. For the biotin-based labels, the yield of the labeling reaction varies considerably, and the most commonly used label, biotin hydrazide, gives the lowest yield. The total tandem mass spectrometry (MS/MS) spectrum counts of modified peptides are similar for all of the biotin-based tags, indicating that factors beyond the labeling efficiency are important in determining the effectiveness of the label. In addition, there is a large variation in the number of spectra obtained for specific, modified peptides depending on the nature of the labeling group. This variation implies that the relative detectability of a particular modification site is highly dependent on the tagging reagent, and more importantly, titration schemes aimed at identifying the most reactive site based on its threshold concentration will be biased by the choice of tagging reagent. The fatty acid hydrazides are somewhat more effective than the biotin-based hydrazides in generating identifiable MS/MS spectra but offer no opportunity for enrichment. For the biotin-based tags, avidin affinity chromatography was used with the tryptic digests, and each tag led to similar enrichment levels.

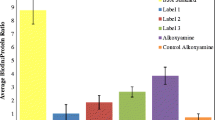
Spectrum counts for HSA peptides modified by acrolein and labeled with different hydrazide tags











Similar content being viewed by others
References
Eal MFLB (2002) Oxidatively modified proteins in aging and disease. Science 32:797–803
Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A (2006) Protein carbonylation, cellular dysfunction, and disease progression. J Cell Mol Med 10:389–406
Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A (2006) Biomarkers of oxidative damage in human disease. Clin Chem 52:601–623
Madian AG, Regnier FE (2010) Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res 9:3766–3780
Suzuki YJ, Carini M, Butterfield DA (2010) Protein carbonylation. Antioxid Redox Signal 12:323–325
Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA (2008) Oxidative stress and covalent modification of protein with bioactive aldehydes. J Biol Chem 283:21837–21841
England K, O’Driscoll C, Cotter TG (2004) Carbonylation of glycolytic proteins is a key response to drug-induced oxidative stress and apoptosis. Cell Death Differ 11:252–260
Linton S, Davies MJ, Dean RT (2001) Protein oxidation and ageing. Exp Gerontol 36:1503–1518
Cabiscol E, Piulats E, Echave P, Herrero E, Ros J (2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem 275:27393–27398
Yan LJ, Sohal RS (1998) Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci U S A 95:12896–12901
Yan LJ, Levine RL, Sohal RS (2000) Effects of aging and hyperoxia on oxidative damage to cytochrome c in the housefly, Musca domestica. Free Radic Biol Med 29:90–97
Yan LJ, Levine RL, Sohal RS (1997) Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci U S A 94:11168–11172
Halliwell B, Whiteman M (2004) Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol 142:231–255
Johansson E, Olsson O, Nyström T (2004) Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 279:22204–22208
Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E, Dukan S (2009) Rules governing selective protein carbonylation. PLoS One. doi:10.1371/journal.pone.0007269
Mirzaei H, Regnier F (2006) Creation of allotypic active sites during oxidative stress. J Proteome Res 5:2159–2168
Mirzaei H, Regnier F (2006) Identification and quantification of protein carbonylation using light and heavy isotope labeled Girard’s P reagent. J Chromatogr A 1134:122–133
Mirzaei H, Regnier F (2005) Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Anal Chem 77:2386–2392
Palmese A, De Rosa C, Marino G, Amoresano A (2011) Dansyl labeling and bidimensional mass spectrometry to investigate protein carbonylation. Rapid Commun Mass Spectrom 25:223–231
Chavez J, Chung W-G, Miranda CL, Singhal M, Stevens JF, Maier CS (2010) Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: protein carbonylation is diminished by ascorbic acid. Chem Res Toxicol 23:37–47
Chavez JD, Bisson WH, Maier CS (2010) A targeted mass spectrometry-based approach for the identification and characterization of proteins containing α-aminoadipic and γ-glutamic semialdehyde residues. Anal Bioanal Chem 398:2905–2914
Aldini G, Gamberoni L, Orioli M, Beretta G, Regazzoni L, Facino RM, Carini M (2006) Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J Mass Spectrom 41:1149–1161
Roe MR, Xie H, Bandhakavi S, Griffin TJ (2007) Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Peptides 79:3747–3756
Guo J, Prokai L (2011) To tag or not to tag: a comparative evaluation of immunoaffinity-labeling and tandem mass spectrometry for the identification and localization of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Proteomics 74:2360–2369
Guo J, Prokai-Tatrai K, Nguyen V, Rauniyar N, Ughy B, Prokai L (2011) Protein targets for carbonylation by 4-hydroxy-2-nonenal in rat liver mitochondria. J Proteomics 74:2370–2379
Rauniyar N, Prokai-Tatrai K, Prokai L (2010) Identification of carbonylation sites in apomyoglobin after exposure to 4-hydroxy-2-nonenal by solid-phase enrichment and liquid chromatography-electrospray ionization tandem mass spectrometry. J Mass Spectrom 45:398–410
Bereman MS, Comins DL, Muddiman DC (2010) Increasing the hydrophobicity and electrospray response of glycans through derivatization with novel cationic hydrazides. Chem Commun 46:237–239
Yoo B-S, Regnier FE (2004) Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis 25:1334–1341
Temple A, Yen T-Y, Gronert S (2006) Identification of specific protein carbonylation sites in model oxidations of human serum albumin. J Am Soc Mass Spectrom 17:1172–1180
Mörtstedt H, Jeppsson MC, Ferrari G, Jönsson BA, Kåredal MH, Lindh CH (2011) Strategy for identification and detection of multiple oxidative modifications within proteins applied on persulfate-oxidized hemoglobin and human serum albumin. Rapid Commun Mass Spectrom 25:327–340
Funk WE, Li H, Iavarone AT, Williams ER, Riby J, Rappaport SM (2010) Enrichment of cysteinyl adducts of human serum albumin. Anal Biochem 400:61–68
Liu Q, Simpson DC, Gronert S (2012) The reactivity of human serum albumin towards trans-4-hydroxy-2-nonenal. J Mass Spectrom 411:411–424
Rossi R, Colombo R, Carini M, Milzani A, Dalle-Donne I (2010) Water-soluble α, β,-unsaturated aldehydes of cigarette smoke induce carbonylation of human serum albumin. Antioxid Redox Signal 12:349–364
Dalle-Donne I, Carini M, Vistoli G, Gamberoni L, Giustarini D, Colombo R, Maffei Facino R, Rossi R, Milzani A, Aldini G (2007) Actin Cys374 as a nucleophilic target of alpha, beta-unsaturated aldehydes. Free Radic Biol Med 42:583–598
Levine RL, Wehr N, Williams JA, Stadtman ER, Shacter E (2000) Determination of carbonyl groups in oxidized proteins. Methods Mol Biol 99:15–24
Washburn MP, Wolters D, Yates JR (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19:242–247
Pang JX, Ginanni N, Dongre AR, Hefta SA, Opiteck GJ (2002) Biomarker discovery in urine by proteomics. J Proteome Res 1:161–169
Gao J, Opiteck GJ, Friedrichs MS, Dongre AR, Hefta SA (2003) Changes in the protein expression of yeast as a function of carbon source. J Proteome Res 2:643–649
Liu H, Sadygov RG, Yates JR (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76:4193–4201
Acknowledgments
We would like to thank Dr. Simpson for his assistance in the implementation of the DNPH protocol. We acknowledge the support of the Virginia Commonwealth University and the National Institute of Health (1R01AG034167).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1.60 kb)
Rights and permissions
About this article
Cite this article
Ugur, Z., Coffey, C.M. & Gronert, S. Comparing the efficiencies of hydrazide labels in the study of protein carbonylation in human serum albumin. Anal Bioanal Chem 404, 1399–1411 (2012). https://doi.org/10.1007/s00216-012-6235-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-012-6235-9