Abstract
Native C-reactive protein (CRP) is composed of five identical subunits arranged in a pentameric structure (pCRP). Binding of pCRP to damaged cell membranes produces a second isoform, modified CRP, which has similar antigenicity to isolated monomeric subunits of CRP (mCRP). Emerging evidence indicates that modified CRP plays a role in inflammation and atherosclerosis, however, there are very few techniques that can distinguish the different isoforms of CRP. Here we show that an RNA aptamer binds specifically to mCRP and not to pCRP. Using this aptamer, we describe a simple, fast, and sensitive assay to detect nanomolar concentrations of mCRP using fluorescence anisotropy. In addition, we show that this aptamer can be used to detect mCRP in polyacrylamide gels and bound to a surface using total internal reflection fluorescence microscopy. The biological activity of the mCRP we prepared by heating pCRP with 0.1% sodium dodecyl sulfate was confirmed by observing binding to the complement protein, C1q. This probe provides an important tool for CRP research and has the potential to improve clinical diagnostics that predict risk for cardiovascular disease.

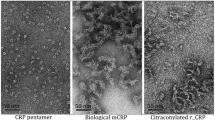
Evidence for mCRP selectivity of aptamer by gel electrophoresis, fluorescence anisotropy, and TIRF microscopy





Similar content being viewed by others
Abbreviations
- CRP:
-
C-reactive protein
- CVD:
-
Cardiovascular disease
- TIRF:
-
Total internal reflection fluorescence
- RNA:
-
Ribonucleic acid
- DNA:
-
Deoxyribonucleic acid
- SELEX:
-
Systemic evolution of ligands by exponential enrichment
- NIR:
-
Near infrared
- DEPC:
-
Diethylpyrocarbonate
- EDTA:
-
Ethylenediaminetetraacetic acid
- ELISA:
-
Enzyme-linked immunosorbant assay
- SDS:
-
Sodium dodecyl sulfate
- HEPES:
-
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- DTT:
-
Dithioreitol
- PBS:
-
Phosphate-buffered saline
- SPR:
-
Surface plasmon resonance
- BSA:
-
Bovine serum albumin
- PAGE:
-
Polyacrylamide gel electrophoresis
- EMSA:
-
Electrophoretic mobility shift assay
References
Black S, Kushner I, Samols D (2004) C-reactive protein. J Biol Chem 279(47):48487–48490
Ji SR, Wu Y, Zhu L, Potempa LA, Sheng FL, Lu W, Zhao J (2007) Cell membranes and liposomes dissociate C-reactive protein (CRP) to form a new, biologically active structural intermediate: mCRP(m). FASEB J 21(1):284–294
Pepys MB, Hirschfield GM (2003) C-reactive protein: a critical update. J Clin Invest 111(12):1805–1812
Thompson D, Pepys MB, Wood SP (1999) The physiological structure of human C-reactive protein and its complex with phosphocholine. Struct Fold Des 7(2):169–177
Volanakis JE (2001) Human C-reactive protein: expression, structure, and function. Mol Immunol 38(2–3):189–197
Potempa LA, Maldonado BA, Laurent P, Zemel ES, Gewurz H (1983) Antigenic, electrophoretic and binding alterations of human C-reactive protein modified selectively in the absence of calcium. Mol Immunol 20(11):1165–1175
Potempa LA, Siegel JN, Fiedel BA, Potempa RT, Gewurz H (1987) Expression, detection and assay of a neoantigen (Neo-Crp) associated with a free, human C-reactive protein subunit. Mol Immunol 24(5):531–541
Potempa LA, Zeller JM, Fiedel BA, Kinoshita CM, Gewurz H (1988) Stimulation of human-neutrophils, monocytes, and platelets by modified C-reactive protein (CRP) expressing a neoantigenic specificity. Inflammation 12(4):391–405
Taylor KE, van den Berg CW (2007) Structural and functional comparison of native pentameric, denatured monomeric and biotinylated C-reactive protein. Immunology 120(3):404–411
Singh SK, Suresh MV, Hammond DJ, Rusinol AE, Potempa LA, Agrawal A (2009) Binding of the monomeric form of C-reactive protein to enzymatically-modified low-density lipoprotein: effects of phosphoethanolamine. Clin Chimica Acta 406(1–2):151–155
Eisenhardt SU, Thiele JR, Bannasch H, Stark GB, Peter K (2009) C-reactive protein: how conformational changes influence inflammatory properties. Cell Cycle 8(23):3885–3892
Khreiss T, Jozsef L, Potempa LA, Filep JG (2004) Conformational rearrangement in C-reactive protein is required for proinflammatory actions on human endothelial cells. Circulation 109(16):2016–2022
Eisenhardt SU, Habersberger J, Murphy A, Chen YC, Woollard KJ, Bassler N, Qian HW, von zur Muhlen C, Hagemeyer CE, Ahrens I, Chin-Dusting J, Bobik A, Peter K (2009) Dissociation of pentameric to monomeric C-reactive protein on activated platelets localizes inflammation to atherosclerotic plaques. Circ Res 105(2):128–155
Gershov D, Kim S, Brot N, Elkon KB (2000) C-reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response: implications for systemic autoimmunity. J Exp Med 192(9):1353–1363
van der Zee PM, Biro E, Trouw LA, Ko Y, de Winter RJ, Hack CE, Sturk A, Nieuwland R (2010) C-reactive protein in myocardial infarction binds to circulating microparticles but is not associated with complement activation. Clin Immunol 135(3):490–495
Volanakis JE, Narkates AJ (1981) Interaction of C-reactive protein with artificial phosphatidylcholine bilayers and complement. J Immunol 126(5):1820–1825
Chang MK, Binder CJ, Torzewski M, Witztum JL (2002) C-reactive protein binds to both oxidized LDL and apoptotic cells through recognition of a common ligand: phosphorylcholine of oxidized phospholipids. Proc Natl Acad Sci USA 99(20):13043–13048
Ying SC, Gewurz H, Kinoshita CM, Potempa LA, Siegel JN (1989) Identification and partial characterization of multiple native and neoantigenic epitopes of human C-reactive protein by using monoclonal antibodies. J Immunol 143(1):221–228
Ji SR, Wu Y, Potempa LA, Liang YH, Zhao J (2006) Effect of modified C-reactive protein on complement activation—a possible complement regulatory role of modified or monomeric C-reactive protein in atherosclerotic lesions. Arterioscl Throm Vas 26(4):935–941
Zouki C, Haas B, Chan JSD, Potempa LA, Filep JG (2001) Loss of pentameric symmetry of C-reactive protein is associated with promotion of neutrophil-endothelial cell adhesion. J Immunol 167(9):5355–5361
Ridker PM, Rifai N, Rose L, Buring JE, Cook NR (2002) Comparison of C-reactive protein and low-density lipoprotein cholesterol levels in the prediction of first cardiovascular events. New Engl J Med 347(20):1557–1565
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287):818–822
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment—RNA ligands to bacteriophage-T4 DNA-polymerase. Science 249(4968):505–510
Tombelli S, Minunni A, Mascini A (2005) Analytical applications of aptamers. Biosens Bioelectron 20(12):2424–2434
Pavlov V, Xiao Y, Shlyahovsky B, Willner I (2004) Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J Am Chem Soc 126(38):11768–11769
Xiao Y, Lubin AA, Heeger AJ, Plaxco KW (2005) Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Edit 44(34):5456–5459
Xiao Y, Piorek BD, Plaxco KW, Heeger AJ (2005) A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J Am Chem Soc 127(51):17990–17991
Minunni M, Tombelli S, Gullotto A, Luzi E, Mascini M (2004) Development of biosensors with aptamers as bio-recognition element: the case of HIV-1 Tat protein. Biosens Bioelectron 20(6):1149–1156
Tombelli S, Minunni A, Luzi E, Mascini M (2005) Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 67(2):135–141
Bini A, Centi S, Tombelli S, Minunni M, Mascini M (2008) Development of an optical RNA-based aptasensor for C-reactive protein. Anal Bioanal Chem 390(4):1077–1086
Huang CJ, Lin HI, Shiesh SC, Lee GB (2010) Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX). Biosens Bioelectron 25(7):1761–1766
Kim SD, Ryu JS, Yi H-K, Kim S-C, Zhang B-T (2004) Construction of C-reactive protein-binding aptamer as a module of the DNA computing system for diagnosing cardiovascular diseases. In: Preliminary proceedings of the tenth international meeting on DNA computing (DNA10):334-343
Stoltenburg R, Reinemann C, Strehlitz B (2005) FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal Bioanal Chem 383(1):83–91
Biro A, Rovo Z, Papp D, Cervenak L, Varga L, Fust G, Thielens NM, Arlaud GJ, Prohaszka Z (2007) Studies on the interactions between C-reactive protein and complement proteins. Immunology 121(1):40–50
Toomre D, Manstein DJ (2001) Lighting up the cell surface with evanescent wave microscopy. Trends Cell Biol 11(7):298–303
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li W, Wang KM, Tan WH, Ma CB, Yang XH (2007) Aptamer-based analysis of angiogenin by fluorescence anisotropy. Analyst 132(2):107–113
Mackiewicz MR, Hodges HL, Reed SM (2010) C-reactive protein induced rearrangement of phosphatidylcholine on nanoparticle mimics of lipoprotein particles. J Phys Chem B 114(16):5556–5562
McCauley TG, Hamaguchi N, Stanton M (2003) Aptamer-based biosensor arrays for detection and quantification of biological macromolecules. Anal Biochem 319(2):244–250
D'Auria S, Barone R, Rossi M, Nucci R, Barone G, Fessas D, Bertoli E, Tanfani F (1997) Effects of temperature and SDS on the structure of beta-glycosidase from the thermophilic archaeon Sulfolobus solfataricus. Biochem J 323:833–840
Agrawal A, Shrive AK, Greenhough TJ, Volanakis JE (2001) Topology and structure of the C1q-binding site on C-reactive protein. J Immunol 166(6):3998–4004
Diehl EE, Haines GK 3rd, Radosevich JA, Potempa LA (2000) Immunohistochemical localization of modified C-reactive protein antigen in normal vascular tissue. Am J Med Sci 319(2):79–83
Wagner M, Weber P, Bruns T, Strauss WSL, Wittig R, Schneckenburger H (2010) Light dose is a limiting factor to maintain cell viability in fluorescence microscopy and single molecule detection. Int J Mol Sci 11(3):956–966
Schwedler SB, Guderian F, Dammrich J, Potempa LA, Wanner C (2003) Tubular staining of modified C-reactive protein in diabetic chronic kidney disease. Nephrol Dial Transpl 18(11):2300–2307
Vaith P, Potempa LA (2000) Deposition of modified or native C-reactive protein in atherosclerotic arteries? Arterioscl Throm Vas 20(4):1173–1174
Hu WP, Hsu HY, Chiou A, Tseng KY, Lin HY, Chang GL, Chen SJ (2006) Immunodetection of pentamer and modified C-reactive protein using surface plasmon resonance biosensing. Biosens Bioelectron 21(8):1631–1637
Singh SK, Suresh MV, Hammond DJ Jr, Rusinol AE, Potempa LA, Agrawal A (2009) Binding of the monomeric form of C-reactive protein to enzymatically-modified low-density lipoprotein: effects of phosphoethanolamine. Clin Chim Acta 406(1–2):151–155
Khreiss T, Jozsef L, Hossain S, Chan JSD, Potempa LA, Filep JG (2002) Loss of pentameric symmetry of C-reactive protein is associated with delayed apoptosis of human neutrophils. J Biol Chem 277(43):40775–40781
Acknowledgements
Support from NIH 1R15GM088960-01 (SMR), NSF CBET-1033161 (SMR), and NSF CBET-1033215 (MKK) is acknowledged. We thank the UCD Biology department for use of the Odyssey imager.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM 1
(PDF 46.7 kb)
Rights and permissions
About this article
Cite this article
Wang, M.S., Black, J.C., Knowles, M.K. et al. C-reactive protein (CRP) aptamer binds to monomeric but not pentameric form of CRP. Anal Bioanal Chem 401, 1309–1318 (2011). https://doi.org/10.1007/s00216-011-5174-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-011-5174-1