Abstract
Hydrophilic interaction chromatography (HILIC) liquid chromatography/mass spectrometry (LC/MS) is appropriate for all native and reductively aminated glycan classes. HILIC carries the advantage that retention times vary predictably according to oligosaccharide composition. Chromatographic conditions are compatible with sensitive and reproducible glycomics analysis of large numbers of samples. The data are extremely useful for quantitative profiling of glycans expressed in biological tissues. With these analytical developments, the rate-limiting factor for widespread use of HILIC LC/MS in glycomics is the analysis of the data. In order to eliminate this problem, a Java-based open source software tool, Manatee, was developed for targeted analysis of HILIC LC/MS glycan datasets. This tool uses user-defined lists of compositions that specify the glycan chemical space in a given biological context. The program accepts high-resolution LC/MS data using the public mzXML format and is capable of processing a large data file in a few minutes on a standard desktop computer. The program allows mining of HILIC LC/MS data with an output compatible with multivariate statistical analysis. It is envisaged that the Manatee tool will complement more computationally intensive LC/MS processing tools based on deconvolution and deisotoping of LC/MS data. The capabilities of the tool were demonstrated using a set of HILIC LC/MS data on organ-specific heparan sulfates.

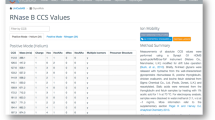
Unsupervised heat map of the 25 most abundant HS compositions among 5 bovine organs with hierarchical clustering of rows and columns






Similar content being viewed by others
References
Wuhrer M, Koeleman CA, Hokke CH, Deelder AM (2005) Protein glycosylation analyzed by normal-phase nano-liquid chromatography–mass spectrometry of glycopeptides. Anal Chem 77:886–894
Bruggink C, Wuhrer M, Koeleman CA, Barreto V, Liu Y, Pohl C, Ingendoh A, Hokke CH, Deelder AM (2005) Oligosaccharide analysis by capillary-scale high-pH anion-exchange chromatography with on-line ion-trap mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 829:136–143
Wuhrer M, Koeleman CAM, Deelder AM, Hokke CN (2004) Normal-phase nanoscale liquid chromatography–mass spectrometry of underivatized oligosaccharides at low-femtomole sensitivity. Anal Chem 76:833–838
Staples GO, Shi X, Zaia J (2010) Extended NS domains reside at the non-reducing end of heparan sulfate chains. J Biol Chem 285:18336–18343
Staples GO, Naimy H, Yin H, Kileen K, Kraiczek K, Costello CE, Zaia J (2010) Improved hydrophilic interaction chromatography LC/MS of heparinoids using a chip with postcolumn makeup flow. Anal Chem 82:516–522
Naimy H, Leymarie N, Zaia J (2010) Screening for anticoagulant heparan sulfate octasaccharides and fine structure characterization using tandem mass spectrometry. Biochemistry 49:3743–3752
Staples G, Bowman M, Costello CE, Hitchcock A, Lau J, Leymarie N, Miller C, Naimy H, Shi X, Zaia J (2009) A chip-based amide-HILIC LC/MS platform for glycosaminoglycan glycomics. Proteomics 9:686–695
Naimy H, Leymarie N, Bowman M, Zaia J (2008) Characterization of heparin oligosaccharides binding specifically to antithrombin III using mass spectrometry. Biochemistry 47:3155–3161
Hitchcock A, Yates KE, Costello C, Zaia J (2008) Comparative glycomics of connective tissue glycosaminoglycans. Proteomics 8:1384–1397
Horn DM, Zubarev RA, McLafferty FW (2000) Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectrom 11:320–332
Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD (2009) Decon2LS: an open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinform 10:87
Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Muller M (2007) SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7:3470–3480
Li XJ, Yi EC, Kemp CJ, Zhang H, Aebersold R (2005) A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol Cell Proteomics 4:1328–1340
Vakhrushev SY, Dadimov D, Peter-Katalinic J (2009) Software platform for high-throughput glycomics. Anal Chem 81:3252–3260
Souady J, Dadimov D, Kirsch S, Bindila L, Peter-Katalinic J, Vakhrushev SY (2010) Software utilities for the interpretation of mass spectrometric data of glycoconjugates: application to glycosphingolipids of human serum. Rapid Commun Mass Spectrom 24:1039–1048
Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol 22:1459–1466
Trans-Proteomic Pipeline, Sashimi Project (2010) Trapper (Mass Hunter converter). Available at http://sourceforge.net/projects/sashimi/files/trapper%20%28MassHunter%20converter%29/
Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang R, Lin C, Chen J, Goodlett D, Whiteaker J, Paulovich A, McIntosh M (2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22:1902–1909
Pacific Northwest National Laboratory (2009) Isotope pattern calculator. Richland. Available at http://omics.pnl.gov/software/IPC.php
Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700–2707
Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman VC, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and computational biology solutions using R and bioconductor, R. Springer, New York, pp 397–420
R Development Core Team (2009) R: a language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. ISBN: 3-900051-07-0. Available at http://www.R-project.org
Esko JD, Selleck SB (2002) Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71:435–471
Bulow HE, Hobert O (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol 22:375–407
Kreuger J, Spillmann D, Li JP, Lindahl U (2006) Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol 174:323–327
Ernst S, Langer R, Cooney CL, Sasisekharan R (1995) Enzymatic degradation of glycosaminoglycans. Crit Rev Biochem Mol Biol 30:387–444
Ahn J, Bones J, Yu YQ, Rudd PM, Gilar M (2010) Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7 microm sorbent. J Chromatogr B Analyt Technol Biomed Life Sci 878:403–408
Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, Lee DM, Rudd PM, Dwek RA (2008) HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem 376:1–12
Domann PJ, Pardos-Pardos AC, Fernandes DL, Spencer DI, Radcliffe CM, Royle L, Dwek RA, Rudd PM (2007) Separation-based glycoprofiling approaches using fluorescent labels. Proteomics 7(Suppl 1):70–76
Hitchcock AM, Yates KE, Shortkroff S, Costello CE, Zaia J (2006) Optimized extraction of glycosaminoglycans from normal and osteoarthritic cartilage for glycomics profiling. Glycobiology 17:25–35
Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M (2000) Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 6:743–750
Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL (2000) Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407:1029–1034
Acknowledgements
Matthew Walsh tested the Manatee program and provided helpful comments on the manuscript. Funding was provided by NIH grants P41RR10888 and R01HL098950 and by an NSF Integrative Graduate Education and Research Traineeship.
This research utilized the Isotope Distribution Calculator developed by the Pacific Northwest National Laboratory, supported by the NIH National Center for Research Resources (Grant RR018522), the W.R. Wiley Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the US Department of Energy's Office of Biological and Environmental Research and located at PNNL), and the National Institute of Allergy and Infectious Diseases (NIH/DHHS through interagency agreement Y1-AI-4894-01). PNNL is operated by Battelle Memorial Institute for the US Department of Energy under contract DE-AC05-76RL0 1830.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the special issue Heparin Characterization with Guest Editor Cynthia K. Larive.
J. M. Dreyfuss, Y. Gindin and C. Jacobs contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2845 kb)
Rights and permissions
About this article
Cite this article
Dreyfuss, J.M., Jacobs, C., Gindin, Y. et al. Targeted analysis of glycomics liquid chromatography/mass spectrometry data. Anal Bioanal Chem 399, 727–735 (2011). https://doi.org/10.1007/s00216-010-4235-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4235-1