Abstract
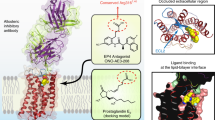
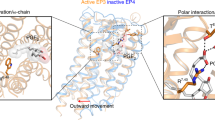
The eight members of the prostanoid receptor family belong to the class A G protein-coupled receptors. We investigated the evolutionary relationship of the eight members by a molecular phylogenetic analysis and found that prostaglandin E2 receptor subtype 2 (EP2) and prostaglandin D2 receptor (DP) were closely related. The structures of the ligands for the two receptors are similar to each other but are distinguished by the exchanged locations of the carbonyl oxygen and the hydroxy group in the cyclopentane ring. The ligand recognition mechanisms of the receptors were examined by an integrated approach using several computational methods, such as amino acid sequence comparison, homology modeling, docking simulation, and molecular dynamics simulation. The results revealed the similar location of the ligand between the two receptors. The common carboxy group of the ligands interacts with the Arg residue on the seventh transmembrane (TM) helix, which is invariant among the prostanoid receptors. EP2 uses a Ser on TM1 to recognize the carbonyl oxygen in the cyclopentane ring of the ligand. The Ser is specifically conserved within EP2. On the other hand, DP uses a Lys on TM2 to recognize the hydroxy group of the ω chain of the ligand. The Lys is also specifically conserved within DP. The interaction network between the D(E)RY motif and TM6 was found in EP2. However, DP lacked this network, due to the mutation in the D(E)RY motif. Based on these observations and the previously published mutational studies on the motif, the possibility of another activation mechanism that does not involve the interaction between the D(E)RY motif and TM6 will be discussed.






Similar content being viewed by others
References
Funk CD (2001) Science 294:1871–1875
Samuelsson B, Morgenstern R, Jakobsson PJ (2007) Pharmacol Rev 59:207–224
Wang MT, Honn KV, Nie D (2007) Cancer Metastasis Rev 26:525–534
Schuligoi R et al (2010) Pharmacology 85:372–382
Jones RL, Giembycz MA, Woodward DF (2009) Br J Pharmacol 158:104–145
Matsuoka T, Narumiya S (2007) Sci World J 7:1329–1347
Hirai H et al (2001) J Exp Med 193:255–261
Hata AN, Breyer RM (2004) Pharmacol Ther 103:147–166
Kedzie KM, Donello JE, Krauss HA, Regan JW, Gil DW (1998) Mol Pharmacol 54:584–590
Chang C, Negishi M, Nishigaki N, Ichikawa A (1997) Biochem J 322:597–601
Stitham J, Stojanovic A, Merenick BL, O’Hara KA, Hwa J (2002) J Biol Chem 278:4250–4257
Funk CD, Furci L, Moran N, Fitzgerald GA (1993) Mol Pharmacol 44:934–939
Neuschäfer-Rube F, Engemaier E, Koch S, Böer U, Püschel GP (2003) Biochem J 371:443–449
Li Y et al (2007) J Am Chem Soc 129:10720–10731
Rosenbaum DM, Rasmussen SG, Kobilka BK (2009) Nature 459:356–363
Scheer A, Fanelli F, Costa T, De Benedetti PG, Cotecchia S (1996) EMBO J 15:3566–3578
Wess J (1998) Pharmacol Ther 80:231–264
Ballesteros J, Palczewski K (2001) Curr Opin Drug Discov Devel 4:561–574
Rosenkilde MM, Kledal TN, Schwartz TW (2005) Mol Pharmacol 68:11–19
Lu ZL, Curtis CA, Jones PG, Pavia J, Hulme EC (1997) Mol Pharmacol 51:234–241
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucleic Acids Res 25:3389–3402
Ballesteros JA, Weinstein H (1995) Methods Neurosci 25:366–428
Katoh K, Misawa K, Kuma K, Miyata T (2002) Nucleic Acids Res 30:3059–3066
Katoh K, Toh H (2008) Brief Bioinform 9:286–298
Saitou N, Nei M (1987) Mol Biol Evol 4:406–425
Felsenstein J (1996) Methods Enzymol 266:418–427
Jones DT, Taylor WR, Thornton JM (1992) Comput Appl Biosci 8:275–282
Felsenstein J (1985) Evolution 39:783–791
Felsenstein J (1993) PHYLIP (phylogeny inference package), version 3.5c. University of Washington, Seattle
Adachi J, Hasegawa M (1996) MOLPHY (programs for molecular phylogenetics), version 2.3b3. Institute of Statistical Mathematics, Tokyo
Halgren TA (1996) J Comput Chem 17:490–519
Labute P (2008) J Comput Chem 29:1693–1698
Wiederstein M, Sippl MJ (2007) Nucleic Acids Res 35:W407–W410
Sippl MJ (1993) Proteins 17:355–362
Goto J, Kataoka R (2008) J Chem Inf Model 48:583–590
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE (2006) In: Proceedings of the ACM/IEEE conference on Supercomputing, Tampa, November 11-17, ACM New York, USA. doi:10.1145/1188455.1188544
Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid B, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallicchio E, Levy RM (2005) J Comput Chem 26:1752–1780
Krautler V (2001) J Comput Chem 22:501–508
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092
Lyne PD, Lamb M, Saeh JC (2006) J Med Chem 49:4805–4808
Toh H, Ichikawa A, Narumiya S (1995) FEBS Lett 361:17–21
Fritze O et al (2003) Proc Natl Acad Sci USA 100:2290–2295
Paila YD, Tiwari S, Chattopadhyay A (2008) Biochim Biophys Acta 1788:295–302
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) Nucleic Acids Res 34:W116–W118
Jaakola V-P, Prilusky J, Sussman JL, Goldman A (2005) Protein Eng Des Sel 18:103–110
Kobayashi T, Ushikubi F, Narumiya S (2000) J Biol Chem 275:24294–24303
Tsuboi K, Sugimoto Y, Ichikawa A (2002) Prostaglandins Other Lipid Mediat 68–69:535–556
Vogel R, Mahalingam M, Lüdeke S, Huber T, Siebert F, Sakmar TP (2008) J Mol Biol 380:648–655
Rovati GE, Capra V, Neubig RR (2007) Mol Pharmacol 71:959–964
Pathe-Neuschäfer-Rube A, Neuschäfer-Rube F, Püschel GP (2005) Biochem J 388:317–324
Ambrosio M, Fanelli F, Brocchetti S, Raimondi F, Mauri M, Rovati GE, Capra V (2010) Cell Mol Life Sci 67:2979–2989
Tusnády GE, Dosztányi Z, Simon I (2005) Bioinformatics 21:1276–1277
Acknowledgments
We thank Drs. Wataru Nemoto, Kentaro Tomii, and Makiko Suwa of CBRC for useful discussions on this work. HD was supported in part by the Global COE program, “an In Silico Medicine”, at Osaka University and Grants-in-Aid (Nos. 20650012 and 19650072) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. HT was partially supported by Targeted Proteins Research Program (TPRP).
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to Professor Akira Imamura on the occasion of his 77th birthday and published as part of the Imamura Festschrift Issue.
Electronic supplementary material
Below is the link to the electronic supplementary material.
214_2011_1034_MOESM2_ESM.eps
Supplement 2 Multiple alignment of prostanoid receptor homologues. The alignment was used to construct the phylogenetic tree shown in Fig. 2. The gi number for each sequence is shown on the left side of the alignment. The information about the source organism and the receptor name are provided in Fig. 2. The D(E)RY motif and the NPxxY motif are indicated by blue rectangles. The second residue in the D(E)RY motif of DP is colored pink (EPS 1,637 kb)
214_2011_1034_MOESM7_ESM.eps
Supplement 7 Time series of the distances between the atom pairs involved in the formation of the intramolecular networks for the EP2 and DP models (EPS 9,880 kb)
Rights and permissions
About this article
Cite this article
Daiyasu, H., Hirokawa, T., Kamiya, N. et al. Computational analysis of ligand recognition mechanisms by prostaglandin E2 (subtype 2) and D2 receptors. Theor Chem Acc 130, 1131–1143 (2011). https://doi.org/10.1007/s00214-011-1034-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00214-011-1034-5