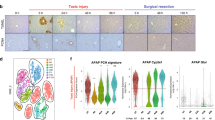
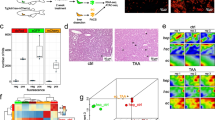
Abstract
Inflammation has been recognized as essential for restorative regeneration. Here, we analyzed the sequential processes during onset of liver injury and subsequent regeneration based on time-resolved transcriptional regulatory networks (TRNs) to understand the relationship between inflammation, mature organ function, and regeneration. Genome-wide expression and TRN analysis were performed time dependently in mouse liver after acute injury by CCl4 (2 h, 8 h, 1, 2, 4, 6, 8, 16 days), as well as lipopolysaccharide (LPS, 24 h) and compared to publicly available data after tunicamycin exposure (mouse, 6 h), hepatocellular carcinoma (HCC, mouse), and human chronic liver disease (non-alcoholic fatty liver, HBV infection and HCC). Spatiotemporal investigation differentiated lobular zones for signaling and transcription factor expression. Acute CCl4 intoxication induced expression of gene clusters enriched for inflammation and stress signaling that peaked between 2 and 24 h, accompanied by a decrease of mature liver functions, particularly metabolic genes. Metabolism decreased not only in pericentral hepatocytes that underwent CCl4-induced necrosis, but extended to the surviving periportal hepatocytes. Proliferation and tissue restorative TRNs occurred only later reaching a maximum at 48 h. The same upstream regulators (e.g. inhibited RXR function) were implicated in increased inflammation and suppressed metabolism. The concomitant inflammation/metabolism TRN occurred similarly after acute LPS and tunicamycin challenges, in chronic mouse models and also in human liver diseases. Downregulation of metabolic genes occurs concomitantly to induce inflammation-associated genes as an early response and appears to be initiated by similar upstream regulators in acute and chronic liver diseases in humans and mice. In the acute setting, proliferation and restorative regeneration associated TRNs peak only later when metabolism is already suppressed.






Similar content being viewed by others
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- NAFLD:
-
Non-alcoholic fatty acid liver disease
- TRN:
-
Transcriptional regulatory networks
- IPA:
-
Ingenuity pathway analysis
- Tm:
-
Tunicamycin
- LPS:
-
Lipopolysaccharide
- HBV:
-
Hepatitis virus type B
References
Arai M, Yokosuka O, Chiba T, Imazeki F, Kato M, Hashida J et al (2003) Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. J Biol Chem 278:29813–29818
Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ (2012) Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. J Biol Chem 287:7345–7356
Braeuning A, Ittrich C, Kohle C, Hailfinger S, Bonin M, Buchmann A et al (2006) Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J 273:5051–5061
Campos G, Schmidt-Heck W, Ghallab A, Rochlitz K, Putter L, Medinas DB et al (2014) The transcription factor CHOP, a central component of the transcriptional regulatory network induced upon CCl4 intoxication in mouse liver, is not a critical mediator of hepatotoxicity. Arch Toxicol 88:1267–1280
Chung H, Hong DP, Jung JY, Kim HJ, Jang KS, Sheen YY et al (2005) Comprehensive analysis of differential gene expression profiles on carbon tetrachloride-induced rat liver injury and regeneration. Toxicol Appl Pharmacol 206:27–42
Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z et al (2010) (2010) Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 468:310–315
Dubois-Pot-Schneider H, Fekir K, Coulouarn C, Glaise D, Aninat C, Jarnouen K et al (2014) Inflammatory cytokines promote the retrodifferentiation of tumor-derived hepatocyte-like cells to progenitor cells. Hepatology 60:2077–2090
Farci P, Diaz G, Chen Z, Govindarajan S, Tice A, Agulto L et al (2010) B cell gene signature with massive intrahepatic production of antibodies to hepatitis B core antigen in hepatitis B virus-associated acute liver failure. Proc Natl Acad Sci USA 107:8766–8771
Forbes SJ, Newsome PN (2016) Liver regeneration—mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 13:473–485
Forbes SJ, Rosenthal N (2014) Preparing the ground for tissue regeneration: from mechanism to therapy. Nat Med 20:857–869
Godoy P, Hewitt NJ, Albrecht U et al (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87(8):1315–1530. https://doi.org/10.1007/s00204-013-1078-5
Godoy P, Schmidt-Heck W, Natarajan K, Lucendo-Villarin B, Szkolnicka D, Asplund A et al (2015) Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J Hepatol 63:934–942
Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL et al (2012) ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Investig 122:4621–4634
Hoehme S, Brulport M, Bauer A, Bedawy E, Schormann W, Hermes M et al (2010) Prediction and validation of cell alignment along microvessels as order principle to restore tissue architecture in liver regeneration. Proc Natl Acad Sci USA 107:10371–10376
Hur KY, So JS, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V et al (2012) IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med 209:307–318
Iracheta-Vellve A, Petrasek J, Gyongyosi B, Satishchandran A, Lowe P, Kodys K et al (2016) Endoplasmic reticulum stress-induced hepatocellular death pathways mediate liver injury and fibrosis via stimulator of interferon genes. J Biol Chem 291:26794–26805
Jansen PL, Ghallab A, Vartak N, Reif R, Schaap FG, Hampe J, Hengstler JG (2017) The ascending pathophysiology of cholestatic liver disease. Hepatology 65(2):722–738. https://doi.org/10.1002/hep.28965
Karin M, Clevers H (2016) Reparative inflammation takes charge of tissue regeneration. Nature 529:307–315
Kim RS, Hasegawa D, Goossens N, Tsuchida T, Athwal V, Sun X et al (2016) The XBP1 arm of the unfolded protein response induces fibrogenic activity in hepatic stellate cells through autophagy. Sci Rep 6:39342
Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV et al (2004) Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology 39:1038–1047
Lazarevich NL, Shavochkina DA, Fleishman DI, Kustova IF, Morozova OV, Chuchuev ES et al (2010) Deregulation of hepatocyte nuclear factor 4 (HNF4)as a marker of epithelial tumors progression. Exp Oncol 32:167–171
Liu Y, Shao M, Wu Y, Yan C, Jiang S, Liu J et al (2015) Role for the endoplasmic reticulum stress sensor IRE1alpha in liver regenerative responses. J Hepatol 62:590–598
Michalopoulos GK (2013) Principles of liver regeneration and growth homeostasis. Compr Physiol 3:485–513
Moylan CA, Pang H, Dellinger A, Suzuki A, Garrett ME, Guy CD et al (2014) Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology 59:471–482
Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S et al (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26:9220–9231
Rashid HO, Yadav RK, Kim HR, Chae HJ (2015) ER stress: autophagy induction, inhibition and selection. Autophagy 11:1956–1977
Santangelo L, Marchetti A, Cicchini C, Conigliaro A, Conti B, Mancone C et al (2011) The stable repression of mesenchymal program is required for hepatocyte identity: a novel role for hepatocyte nuclear factor 4alpha. Hepatology 53:2063–2074
Wang B, Cai SR, Gao C, Sladek FM, Ponder KP (2001) Lipopolysaccharide results in a marked decrease in hepatocyte nuclear factor 4 alpha in rat liver. Hepatology 34:979–989
White P, Brestelli JE, Kaestner KH, Greenbaum LE (2005) Identification of transcriptional networks during liver regeneration. J Biol Chem 280:3715–3722
Young MB, DiSilvestro MR, Sendera TJ, Freund J, Kriete A, Magnuson SR (2003) Analysis of gene expression in carbon tetrachloride-treated rat livers using a novel bioarray technology. Pharmacogenom J 3:41–52
Zidek N, Hellmann J, Kramer PJ, Hewitt PG (2007) Acute hepatotoxicity: a predictive model based on focused illumina microarrays. Toxicol Sci 99:289–302
Acknowledgements
We thank Ms. Katharina Rochlitz, Ms. Brigitte Begher-Tibbe, and Ms. Georgia Günther (Leibniz Research Centre for Working Environment and Human Factors, Technical University of Dortmund, Germany) for excellent technical assistance. This work has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 681002 (EU-ToxRisk), from TransQST (No. 116030), LivSysTransfer (BMBF, 031L0119) with contributions from StemCellNet (BMBF, 01EK1604A), LiSyM (BMBF, 031Loo45), and InnoSysTox (BMBF/EU, 031L0021A) We thank the Permanent Senate Commission on Food Safety (SKLM) for valuable discussion.
Funding
This study was supported by the European Union Seventh Framework Programme (FP7) Health projects DETECTIVE (EU-project FP7-Health Grant agreement no. 266838) and NOTOX (EU-project FP7-Health Grant agreement no. 267038) (JGH), the DFG WISP (GO 1987/2-1) (PG), the BMBF (German Federal Ministry of Education and Research) project Virtual Liver (0313854) (JGH), FONDECYT no. 1140549, Millennium Institute no. P09-015-F and FONDAP 15150012 (CH).
Author information
Authors and Affiliations
Contributions
GC, AW, AG, LP mouse experiments with CCl4, Tm and LPS challenge; GC, AW, LP western blot analyses; GC histological analysis; WS-H, PG, JS and AS gene array analysis and bioinformatics; AB and MS zonated gene array analysis; GC, DG real-time PCR analysis; GC, JGH and PG drafting of manuscript, acquisition of funding; GC, JGH and PG Study concept and design, critical review of manuscript, study supervision; KE, CC, RM, RR, RG, CV, CH, MS, TSW, JLS, DCH and DD study concept and design, critical revision of manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Campos, G., Schmidt-Heck, W., De Smedt, J. et al. Inflammation-associated suppression of metabolic gene networks in acute and chronic liver disease. Arch Toxicol 94, 205–217 (2020). https://doi.org/10.1007/s00204-019-02630-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-019-02630-3