Abstract
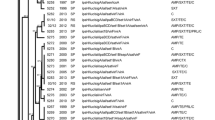
Shigellosis is the main cause of food and waterborne diarrhea and is an emerging threat to human health. The current study characterized the indigenous multidrug-resistant Shigella flexneri serotypes for their plasmid profiles and genetic diversity, to characterize the plasmid evolutionary patterns and distribution. In total, 199 identified S. flexneri isolates belonging to six different serotypes were analyzed for plasmid profiling, followed by an analysis of whole genome sequencing. All isolates of S. flexneri resistant to antibiotics harbored multiple copies of plasmids with sizes ranging from 1.25 kbp to 9.4 kbp. These isolates were clustered into 22 distinct plasmid patterns, labeled as p1–p22. Among these, p1 (24%) and p10 (13%) were the predominant plasmid profiles. All S. flexneri strains were grouped into 12 clades with a 75% similarity level. Also, a significant association was observed among the plasmid patterns, p23 and p17 with the drug-resistant patterns AMC, SXT, C (19.5%) and OFX, AMC, NA, CIP (13.5%), respectively. Moreover, the most widespread plasmid patterns p4, p10, and p1 showed a significant association with the serotypes 1b (29.16%), 2b (36%), and 7a (100%), respectively. After plasmid sequence assembly and annotation analysis, a variety of small plasmids that vary in size from 973 to 6200 bp were discovered. Many of these plasmids displayed high homology and coverage with plasmids from non-S. flexneri. Several novel plasmids of small size were discovered in multidrug-resistant S. flexneri. The data also showed that plasmid profile analysis is more consistent than antibiotic susceptibility pattern analysis for identifying epidemic strains of S. flexneri isolated in Pakistan.

Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
Agtini MD, Soeharno R, Lesmana M et al (2005) The burden of diarrhoea, shigellosis, and cholera in North Jakarta, Indonesia: findings from 24 months surveillance. BMC Infect Dis 5:89
Ahmed B, Shakoori FR, Ali SS et al (2009) antimicrobial resistance and plasmid profile analysis of clinically isolated Shigella dysenteriae in Azad Kashmir, Pakistan. Pak J Zool 41(6):495–503
Aziz KMA, Wells JG, Boyce JM et al (1982) Patterns of Shigella infection in families in Rural Bangladesh. Am J Trop Med Hyg 31:1015–1020
Banner DW, Kokkinidis M, Tsernoglou D (1987) Structure of the ColE1 rop protein at 1.7A resolution. J Mol Biol 196:657–675
Boyd AC, Archer JA, Sherratt DJ (1989) Characterization of the ColE1 mobilization region and its protein products. Mol Gen Genet 217:488–498
Burian J, Ausió J, Phipps B et al (2003) Hexamerization of RepA from the Escherichia coli plasmid pKL1. Biochemistry 42:10282–10287
Craun GF, Calderon RL, Craun MF (2005) Outbreaks associated with recreational water in the United States. Int J Environ Health Res 15:243–262
Davies J, Wright GD (1997) Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 5:234–240
Deatherage DE, Barrick JE (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188
Dutta S, Rajendran K, Roy S et al (2002) Shifting serotypes, plasmid profile analysis and antimicrobial resistance pattern of shigellae strains isolated from Kolkata, India during 1995–2000. Epidemiol Infect 129:235–243
Farshad S, Sheikhi R, Japoni A et al (2006) Characterization of Shigella Strains in Iran by plasmid profile analysis and PCR amplification of ipa Genes. J Clin Microbiol 44:2879–2883
Kamada K, Hanaoka F, Burley SK (2003) Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell 11:875–884
Kim C, Mobashery S (2005) Phosphoryl transfer by aminoglycoside 3′-phosphotransferases and manifestation of antibiotic resistance. Bioorg Chem 33:149–158
Kotloff KL, Winickoff JP, Ivanoff B et al (1999) Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 77:651–666
Lane BR, Ast JC, Hossler PA et al (1997) Dihydropteroate synthase polymorphisms in Pneumocystis carinii. J Infect Dis 175:482–485
Murray GL, Attridge SR, Morona R (2005) Inducible serum resistance in Salmonella typhimurium is dependent on wzzfepE-regulated very long O antigen chains. Microbes Infect 7:1296–1304
Nandy S, Mitra U, Rajendran K, Dutta P, Dutta S (2010) Subtype prevalence, plasmid profiles and growing fluoroquinolone resistance in Shigella from Kolkata India (2001–2007): a hospital-based study. Trop Med Int Heal 15:1499–1507
Nisa I, Qasim M, Driessen A et al (2020) Molecular epidemiology of Shigella flexneri isolated from pediatrics in a diarrhea-endemic area of Khyber Pakhtunkhwa, Pakistan. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-020-03811-0
Nisa I, Qasim M, Yasin N et al (2020b) Shigella flexneri: an emerging pathogen. Folia Microbiol (praha). https://doi.org/10.1007/s12223-020-00773-w
Nisa I, Qasim M, Driessen A et al (2021) Prevalence and associated risk factors of Shigella flexneri isolated from drinking water and retail raw foods in Peshawar. Pakistan J Food Sci 86:2579–2589
Nisa I, Haroon M, Driessen A et al (2022) Antimicrobial resistance of Shigella flexneri in Pakistani pediatric population reveals an increased trend of third-generation cephalosporin resistance. Curr Microbiol 79:1–10
Nurizzo D, Shewry SC, Perlin MH et al (2003) The crystal structure of aminoglycoside-3’-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J Mol Biol 327:491–506
Ozenberger BA, Nahlik MS, McIntosh MA (1987) Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J Bacteriol 169:3638–3646
Sakhaei A, Savari M, Shokoohizadeh L et al (2015) Characterization of Shigella strains by plasmid profile analysis and antibiotic susceptibility patterns in a pediatric hospital in Ahvaz. Int J Enteric Pathog 3:29924
Sharma A, Kumar S, Divya S (2009) Phenotypic and genotypic characterization of Shigella spp. with reference to its virulence genes and antibiogram analysis from river Narmada. Indian J Microbiol 49:259–265
Szakál DD, Schneider G, Pál T (2003) A colony blot immune assay to identify enteroinvasive Escherichia coli and Shigella in stool samples. Diagn Microbiol Infect Dis 45:165–171
Tacket CO, Shahid N, Huq MI, Alim ARMA, Cohen ML (1984) Usefulness of plasmid profiles for differentiation of Shigella isolates in Bangladesh enteric diseases branch, centers for disease control. J Clin Microbiol 20:300–301
Talukder KA, Islam Z, Islam MA et al (2003) Phenotypic and genotypic characterization of provisional serotype Shigella flexneri 1c and clonal relationships with 1a and 1b strains isolated in Bangladesh. J Clin Microbiol 41:110–117
Tohyama H, Okami Y, Umezawa H (1987) Nucleotide sequence of the streptomycinphosphotransferase and amidinotransferase genes from Streptomyces griseus. Nucleic Acids Res 15:1819–1833
Venkatesan MM, Goldberg MB, Rose DJ et al (2001) Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun 69:3271–3285
Wei Y, Murphy ER (2016) Shigella iron acquisition systems and their regulation. Front Cell Infect Microbiol 6:18
Zhang S, Meyer R (1997) The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol Microbiol 25:509–516
Acknowledgements
The authors would like to acknowledge the staff of the Department of Molecular Microbiology Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, The Netherland, Veterinary Research Institute Peshawar, Lady Reading Hospital Peshawar, Pakistan and Kohat University of Science and Technology, Kohat, for their excellent technical assistance. This work was supported by grants from the Higher Education Commission of Pakistan (FTO number 0341-3970-2018). In addition, the current work is a part of Iqbal Nisa’s PhD thesis/dissertation.
Funding
This work was supported by grants from the Higher Education Commission of Pakistan (FTO number 0341-3970-2018).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by IN and analysis was performed by MQ, AD, JN, HR, and JM. The first draft of the manuscript was written by IN and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no relevant financial or non-financial interests to disclose.
Ethical approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000, approved by the Kohat University of Science and Technology, Kohat research and ethical committee.
Consent to participate
Informed consent was obtained from all patients/guardians for being included in the study.
Consent for publication
Not applicable.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.











Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nisa, I., Driessen, A., Nijland, J. et al. Novel plasmids in multidrug-resistant Shigella flexneri serotypes from Pakistan. Arch Microbiol 205, 175 (2023). https://doi.org/10.1007/s00203-023-03523-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03523-x