Abstract
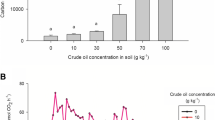
The bacterial community composition in soil sediments changes with respect to nutrient concentrations and environmental conditions. Reports on the correlation between bacterial populations and inorganic nutrient concentrations in oily sediments are limited. The present time series study reports the prevalence of specific hydrocarbon-degrading bacterial communities in nutrient-treated oily sludge microcosms. The hydrocarbon degradation was maximum at 625 µg nitrogen (N) and 62.5 µg phosphorus (P)/g sludge sediment. The 16S rRNA gene-based DGGE analyses revealed noticeable changes in bacterial community composition with time and levels of nutrient treatment. BLASTn analysis of the 16S rRNA gene clone sequence showed the abundance of γ-Proteobacteria (44%), α-Proteobacteria (16%), β-Proteobacteria (10%), CFB (4%), and unidentified bacterial clones (26%). The catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O) gene clones were affiliated to the genus Sphingomonas, highlighting the vital role of Sphingomonas in aromatic hydrocarbon degradation. The quantity of the 16S rRNA gene and the alkane hydroxylase (alkB) gene reached maximum levels in extended duration microcosms treated with 625 µg N and 62.5 µg P/g sludge sediment. In contrast, the C12O gene reached its highest abundance at a low N concentration.





Similar content being viewed by others
References
Abbasian F, Palanisami T, Megharaj M, Naidu R, Lockington R, Ramadass K (2016) Microbial diversity and hydrocarbon degrading gene capacity of a crude oil field soil as determined by metagenomics analysis. Biotechnol Prog 32(3):638–648
Al-Hawash AB, Dragh MA, Li S, Alhujaily A, Abbood HA, Zhang X, Ma F (2018) Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt J Aquat Res 44(2):71–76
Atlas RM (1981) Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev 45(1):180–209
Bacosa H, Suto K, Inoue C (2010) Prefential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int Biodeter Biodegr 64(8):702–710
Beolchini F, Roccheti L, Regoli F, DellʹAnno A (2010) Bioremediation of marine sediments contaminated by hydrocarbons: experimental analysis and kinetic modelling. J Hazard Mater 182(1–3):403–407
Bordenave S, Goñi-Urriza MS, Caumette P, Duran R (2007) Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat. Appl Environ Microbiol 73(19):6089–6097
Cerqueira VS, Hollenbach EB, Maboni F, Vainstein MH, Camargo FAO, Peralba MCR, Bento FM (2011) Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol 102(23):11003–11010
Chettri B, Mukherjee A, Langpoklakpam JS, Chattopadhyay D, Singh AK (2016) Kinetics of nutrient enhanced crude oil degradation by Pseudomonas aeruginosa AKS1 and Bacillus sp. AKS2 isolated from Guwahati refinery, India. Environ Poll 216:548–558
Chettri B, Singha NA, Mukherjee A, Rai AN, Chattopadhyay DJ, Singh AK (2019) Hydrocarbon degradation potential and competitive persistence of hydrocarbonoclastic bacterium Acinetobacter pitti strain ABC. Arch Microbiol 201(8):1129–1140
Chettri B, Singha NA, Singh AK (2021) Efficiency and kinetics of Assam crude oil degradation by Pseudomonas aeruginosa and Bacillus sp. Arch Microbiol 203(9):5793–5803
Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN (2007) Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ Microbiol 9(1):177–186
Cydzik-Kwiatkowska A, Zielińska M (2016) Bacterial communities in full-scale wastewater treatment systems. World J Microbiol Biotechnol 32(4):1–8
Da Silva VL, Alves FC, De Franca FP (2012) A review of the technological solutions for the treatment of oily sludges from petroleum refineries. Waste Manage Res 30:1016–1030
Das K, Mukherjee AK (2007) Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour Technol 98(7):1339–1345
Das R, Kazy SK (2014) Microbial diversity, community composition and metabolic potential in hydrocarbon contaminated oily sludge: prospects for in situ bioremediation. Environ Sci Pollut Res Int 21(12):7369–7389
Falkowski PG, Fenchel T, Delong EF (2008) The microbial engines that drive earth’s biogeochemical cycles. Science 320(5879):1034–1039
Gao W, Yin X, Mi T, Zhang Y, Lin F, Han B, Zhao X, Luan X, Cui Z, Zhang L (2018) Microbial diversity and ecotoxicity of sediments 3 years after the Jiaozhou Bay Oil Spill. AMB Expr 8(1):1–10
Head IM, Jones DM, Röling WFM (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4(3):173–182
Khomarbaghi Z, Shavandi M, Amoozegar MA, Dastgheib M (2019) Bacterial community dynamics during bioremediation of alkane- and PAHs-contaminated soil of Siri Island, Persian Gulf: a microcosm study. Int J Environ Sci Technol 16:7849–7860
Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M (2011) Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol 77(22):7962–7974
Kumar B, Rajmohan B (2013) Petroleum oily sludge and the prospects of microwave for its remediation. Int J Eng Res Technol 2(11):359–370
Ladino-Orjuela G, Gomes E, da Silva R, Salt C, Parsons JR (2016) Metabolic pathways for degradation of aromatic hydrocarbons by bacteria. Rev Environ Contam Toxicol 237:105–121
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackenbrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley, New York, pp 115–148
Ławniczak L, Wozniak-Karczewska M, Loibner AP, Heipieper HJ, Chrzanowski L (2020) Microbial degradation of hydrocarbons—basic principles for bioremediation: a review. Molecules 25(4):856
Liu W, Luo Y, Teng Y, Li Z, Ma LQ (2010) Bioremediation of oily sludge-contaminated soil by stimulating indigenous microbes. Environ Geochem Health 32(1):23–29
Liu Q, Tang J, Bai Z, Hecker M, Giesy JP (2015) Distribution of petroleum degrading genes and factor analysis of petroleum contaminated soil from the Dagang Oilfield, China. Sci Rep 5:11068
Looper JK, Cotto A, Kim BY, Lee MK, Liles MR, Ni Chadhain SM, Son A (2013) Microbial community analysis of Deepwater Horizon oil-spill impacted sites along the Gulf coast using functional and phylogenetic markers. Environ Sci Process Impacts 15(11):2068–2079
McKew BA, Coulon BA, Yakimov F, Denaro R, Genovese M, Smith CJ, Osborn AM, Timmis KN, McGenity TJ (2007) Efficacy of intervention strategies for bioremediation of crude oil in marine systems and effects on indigenous hydrocarbonoclastic bacteria. Environ Microbiol 9(6):1562–1571
Mukherjee A, Chettri B, Langpoklakpam JS, Basak P, Prasad A, Mukherjee AK, Bhattacharyya M, Singh AK, Chattopadhyay D (2017) Bioinformatic approaches including predictive metagenomic profiling reveal characteristics of bacterial response to petroleum hydrocarbon contamination in diverse environments. Sci Rep 7(1):1–22. https://doi.org/10.1038/s41598-017-01126-3
Muyzer G, Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59(3):695–700
Neihsial R, Singha NA, Singh AK (2022) Taxonomic diversity and predictive metabolic functions of a heavy metal tolerant multiple azo dye degrading bacterial consortium from textile effluents. Int Biodeter Biodegr 171:105421
Nie Y, Chi CQ, Fang H, Liang JL, Lu SL, Lai GL, Tang YQ, Wu XL (2014) Diverse alkane hydroxylase genes in microorganisms and environments. Sci Rep 4(1):1–11
Nilsen OG, Haugen OA, Zahlsen K, Halgunset J, Helseth A, Aarset H, Eide I (1988) Toxicity of n-C9 to n-C13 alkanes in the rat on short term inhalation. Pharmacol Toxicol 62(5):259–266
Röling WF, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJ, Head IM (2002) Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68(11):5537–5548
Roy AS, Baruah R, Borah M, Singh AK, Deka Boruah HP, Saikia N, Deka M, Dutta N, Chandra Bora T (2014) Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int Biodeter Biodegr 94:79–89
Roy A, Dutta A, Pal S, Gupta A, Sarkar J, Chatterjee A, Saha A, Sarkar P, Sar P, Kazy SK (2018) Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour Technol 253:22–32
Sei K, Asano K, Tateishi N, Mori K, Ike M, Fujita M (1999) Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways. J Biosci Bioeng 88(5):542–550
Sei K, Sugimoto Y, Mori K, Maki H, Kohno T (2003) Monitoring of alkane-degrading bacteria in a sea-water microcosm during crude oil degradation by polymerase chain reaction based on alkane-catabolic genes. Environ Microbiol 5(6):517–522
Shahi A, Aydin S, Ince B, Ince O (2016) Reconstruction of bacterial community structure and variation for enhanced petroleum hydrocarbons degradation through biostimulation of oil contaminated soil. Chem Eng J 306:60–66
Singh AK, Sherry A, Gray ND, Jones MD, Röling WFM, Head IM (2011) How specific microbial communities benefit the oil industry: dynamics of Alcanivorax spp. in oil-contaminated intertidal beach sediments undergoing bioremediation. In: Whitby C, Skovhus TL (eds) Applied microbiology and molecular biology in oilfield systems. Springer, Dordrecht, Netherlands, pp 199–209
Singh AK, Sherry A, Gray ND, Jones DM, Bowler BF, Head IM (2014) Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front Microbiol 5:160
Swann HE, Kwon BK, Hogan GK, Snellings WM (1974) Acute inhalation toxicology of volatile hydrocarbons. Am Ind Hyg Assoc J 35(9):511–518
Tao K, Zhang Z, Chen X, Liu X, Hu X, Yuan X (2019) Response of soil bacterial community to bioaugmentation with a plant residue-immobilized bacterial consortium for crude oil removal. Chemosphere 222:831–838
Turner S, Pryer KM, Miao VP, Palmer JD (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46(4):327–338
Varjani SJ (2017) Microbial degradation of petroleum hydrocarbons. Biores Technol 223:277–286
Varjani SJ, Gnansounou E, Pandey A (2017) Comprehensive review on toxicity of persistent organic pollutants from petroleum refinery waste and their degradation by microorganisms. Chemosphere 188:280–291
Varjani S, Pandey A, Upasani VN (2020) Oilfield waste treatment using novel hydrocarbon utilizing bacterial consortium—a microcosm approach. Sci Total Environ 745:141043
Verma S, Bhargava R, Pruthi V (2006) Oily sludge degradation by bacteria from Ankleshwar, India. Int Biodeter Biodegr 57(4):207–213
Viggor S, Juhanson J, Joesaar M, Mitt M, Truu J, Vedler E, Heinaru A (2013) Dynamic changes in the structure of microbial communities in Baltic Sea coastal seawater microcosms modified by crude oil, shale oil or diesel fuel. Microbiol Res 168(7):415–427
Wang M, Sha C, Wu J, Su J, Wu J, Wang Q, Tan J, Huang S (2021) Bacterial community response to petroleum contamination in brackish tidal marsh sediments in the Yangtze River Estuary, China. J Environ Sci 99:160–167
Wasmund K, Burns KA, Kurtböke DI, Bourne DG (2009) Novel alkane hydroxylase gene (alkB) diversity in sediments associated with hydrocarbon seeps in the Timor Sea, Australia. Appl Environ Microbiol 75(23):7391–7398
Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, Cummings TA, Beers AR, Knight R, Angenent LT (2011) Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci USA 108(10):4158–4163
Zafra G, Absalón AE, Cortés-Espinosa DV (2015) Morphological changes and growth of filamentous fungi in the presence of high concentrations of PAHs. Braz J Microbiol 46:937–941
Acknowledgements
The authors are thankful to the Department of Biochemistry, North Eastern Hill University, for providing the research facilities. Fellowship grant from DST INSPIRE to Bobby Chettri (IF10272) is gratefully acknowledged.
Funding
This research was supported by a grant from DBT, Govt. of India (BT/306/NE/TBP/2012).
Author information
Authors and Affiliations
Contributions
AKS and CA developed the concept of the experiments. BC conducted the experiments. All authors contributed to data analysis and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chettri, B., Akoijam, C. & Singh, A.K. Dynamics and prevalence of specific hydrocarbonoclastic bacterial population with respect to nutrient treatment levels in crude oil sludge. Arch Microbiol 204, 708 (2022). https://doi.org/10.1007/s00203-022-03323-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03323-9