Abstract
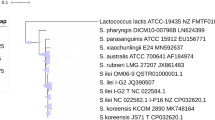
Among the species of genus Streptococcus two members namely Streptococcus ratti and Streptococcus ursoris were isolated from oral cavity of rat and bear, respectively. Type strain of these members shows a 16S rRNA gene sequence similarity of 98.9%. Based on systematic phylo-taxonogenomics investigations, we could deduce the taxonomic assignment of the members of these species. Genome similarity assessment among the type strain of these members using average nucleotide identity (orthoANI and fastANI), digital DNA–DNA hybridization and average amino acid identity (AAI) were 98.5, 98.3, 88, and 98.3% respectively. All these values exceed the species delineation cutoffs suggesting a unified species. Phylogenetic tree obtained using 16S rRNA gene sequence also indicates the monophyletic nature of the member strains. Such monophyletic taxonomic positioning of the strains was further complemented with the whole genome-based phylogenomic tree. Based on these evidences, we propose S. ursoris should be reclassified as a later heterotypic synonym of S. ratti.


Similar content being viewed by others
Abbreviations
- CSI:
-
Conserved sequence indels
- ANI:
-
Average Nucleotide Identity
- dDDH:
-
Digital DNA–DNA hybridization
- ML:
-
Maximum likelihood
- GS:
-
Genomospecies
References
Andrewes F, Horder T (1906) A study of the streptococci pathogenic for man. The Lancet 168:775–783
Asnicar F et al (2020) Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat Commun 11:1–10. https://doi.org/10.1038/s41467-020-16366-7
Auch AF, Klenk H-P, Göker M (2010) Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2:142–148
Bansal K, Kumar S, Kaur A, Singh A, Patil PB (2021) Deep phylo-taxono genomics reveals Xylella as a variant lineage of plant associated Xanthomonas and supports their taxonomic reunification along with Stenotrophomonas and Pseudoxanthomonas. Genomics 113:3989–4003
Bridge P, Sneath P (1982) Streptococcus gallinarum sp. nov. and Streptococcus oralis sp. nov. Int J Syst Evol Microbiol 32:410–415
Chun J et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Coykendall A (1974) Four types of Streptococcus mutans based on their genetic, antigenic and biochemical characteristics. Microbiology 83:327–338
Coykendall AL (1977) Proposal to elevate the subspecies of Streptococcus mutans to species status, based on their molecular composition. Int J Syst Evol Microbiol 27:26–30
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Facklam RR (1977) Physiological differentiation of viridans streptococci. J Clin Microbiol 5:184–201
Fazili T et al (2017) Streptococcus anginosus group bacterial infections. Am J Med Sci 354:257–261
Handley P, Coykendall A, Beighton D, Hardie J, Whiley R (1991) Streptococcus crista sp. nov., a viridans streptococcus with tufted fibrils, isolated from the human oral cavity and throat. Int J Syst Evol Microbiol 41:543–547
Jablon JM, Zinner DD (1966) Differentiation of cariogenic streptococci by fluorescent antibody. J Bacteriol 92:1590–1596
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:1–8
Kawamura Y et al (1998) Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Int J Syst Evol Microbiol 48:921–927
Kumar S, Bansal K, Sethi SK (2022) Phylo-taxonogenomic reconciliations of genus Streptococcus reveals the existence of sixteen novel genomospecies. Res Square https://doi.org/10.21203/rs.3.rs-1416559/v1
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kumar S, Bansal K, Patil PP, Patil PB (2019) Phylogenomics insights into order and families of Lysobacterales. Access Microbiol. https://doi.org/10.1099/acmi.0.000015
Lee I, Chalita M, Ha S-M, Na S-I, Yoon S-H, Chun J (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057
Meehan CJ, Barco RA, Loh Y-HE, Cogneau S, Rigouts L (2021) Reconstituting the genus Mycobacterium. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.004922
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807
Orla-Jensen S (1919) The lactic acid bacteria. Høst
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Patel S, Gupta RS (2018) Robust demarcation of fourteen different species groups within the genus Streptococcus based on genome-based phylogenies and molecular signatures. Infect Genet Evol 66:130–151
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci 106:19126–19131
Rosenbach AJF (1884) Mikro-organismen bei den Wund-infections-krankheiten des Menschen. JF Bergmann
Shinozaki-Kuwahara N, Takada K, Hirasawa M (2011) Streptococcus ursoris sp. nov., isolated from the oral cavities of bears. Int J Syst Evol Microbiol 61:40–44
Stackebrandt E (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Trüper HG, De’clari L (1997) Taxonomic note: necessary correction of specific epithets formed as substantives (nouns)“in apposition.” Int J Syst Evol Microbiol 47:908–909
Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Zinner D, Jablon J, Aran A, Saslaw M, Fitzgerald R (1966) Comparative pathogenicity of streptococci of human origin in hamster caries. Arch Oral Biol 11:1419-IN1447
Zinner DD, Aran AP, Saslaw MS, JablonFitzgerald JMRJ (1967) Induction of dental caries in gnotobiotic rats by streptococci of human origin. Nature 213:200–201
Acknowledgements
Authors convey a special thanks to Dr. Prabhu Patil, CSIR-IMTECH, Chandigarh for their immense support throughout the whole study. Authors also convey their gratitude towards the critical inputs from Dr. Kanika Bansal, CSIR-IMTECH, Chandigarh for her suggestion on the manuscripts.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
SK have performed the analysis and drafted manuscript with the inputs from SKS. SKS have supervised the study by performing data correction, formal analysis and drafting of methodology.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of interest.
Ethical approval
This is an observational study which does not require any ethical approval.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, S., Sethi, S.K. Genome-based reclassification of Streptococcus ursoris as a later heterotypic synonym of Streptococcus ratti. Arch Microbiol 204, 405 (2022). https://doi.org/10.1007/s00203-022-03012-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03012-7