Abstract
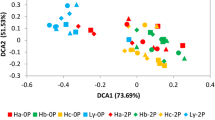
Global warming is an indisputable fact. However, the effect of warming on the rhizosphere bacterial community of crops is not well understood. Therefore, we carried out pot experiments with three rice (Oryza sativa L.) varieties in black soil across three climatic regions of northeast China to simulate temperature change, and analyzed the response of the rhizosphere bacterial community to different temperatures. Results showed that climate had stronger effects on rhizosphere bacterial communities than rice variety. The rhizosphere bacterial diversity differed significantly among the three climatic regions and positively correlated with the mean daily average temperature (MAveT), mean daily maximum temperature (MMaxT), and mean daily minimum temperature (MMinT), and negatively correlated with the daily temperature range (DTR). Principal co-ordinate analysis revealed that bulk soil bacterial communities maintained a high similarity across the three climatic regions, while rhizosphere bacterial communities notably varied. This change was significantly correlated with MAveT, MMaxT, MMinT, and DTR. Compared with bulk soil, Proteobacteria and Bacteroidetes were enriched in the rhizosphere, while Actinobacteria was depleted. Moreover, these changes were strengthened by increasing the temperature and decreasing DTR. Additionally, correlation analysis revealed that changes in rhizosphere bacterial communities were closely related to the formation of rice yields. Our study revealed that the increasing temperature indirectly reshapes the rhizosphere bacterial community that may promote rice production in areas with lower temperatures.






Similar content being viewed by others
References
Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL (2018) The soil-borne legacy. Cell 172:1178–1180. https://doi.org/10.1016/j.cell.2018.02.024
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. ICWSM https://doi.org/10.13140/2.1.1341.1520
Bhattacharyya P, Roy KS, Das M, Ray S, Balachandar D, Karthikeyan S, Nayak AK, Mohapatra T (2016) Elucidation of rice rhizosphere metagenome in relation to methane and nitrogen metabolism under elevated carbon dioxide and temperature using whole genome metagenomic approach. Sci Total Environ 542:886–898. https://doi.org/10.1016/j.scitotenv.2015.10.154
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. https://doi.org/10.1038/nmeth.2276
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E et al (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. https://doi.org/10.1038/nature11336
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth0510-335
Chen Q, Niu B, Hu Y, Luo T, Zhang G (2020) Warming and increased precipitation indirectly affect the composition and turnover of labile-fraction soil organic matter by directly affecting vegetation and microorganisms. Sci Total Environ 714:136787. https://doi.org/10.1016/j.scitotenv.2020.136787
Chen S, Waghmode TR, Sun R, Kuramae EE, Hu C, Liu B (2019) Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7:136. https://doi.org/10.1186/s40168-019-0750-2
Das S, Bhattacharyya P, Adhya TK (2010) Impact of elevated CO2, flooding, and temperature interaction on heterotrophic nitrogen fixation in tropical rice soils. Biol Fertil Soils 47:25–30. https://doi.org/10.1007/s00374-010-0496-2
Ding L, Cui H, Nie S, Long X, Duan G, Zhu Y (2019) Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol Ecol 95:fiz040. https://doi.org/10.1093/femsec/fiz040
Dresbøll DB, Thorup-Kristensen K (2012) Spatial variation in root system activity of tomato (Solanum lycopersicum L.) in response to short and long-term waterlogging as determined by 15N uptake. Plant Soil 357:161–172. https://doi.org/10.1007/s11104-012-1135-5
Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:911–920. https://doi.org/10.1073/pnas.1414592112
Epelde L, Becerril JM, Barrutia O, Gonzalez-Oreja JA, Garbisu C (2010) Interactions between plant and rhizosphere microbial communities in a metalliferous soil. Environ Pollut 158:1576–1583. https://doi.org/10.1016/j.envpol.2009.12.013
Fan K, Delgado-Baquerizo M, Guo X, Wang D, Wu Y, Zhu M, Yu W, Yao H, Zhu YG, Chu H (2019) Suppressed N fixation and diazotrophs after four decades of fertilization. Microbiome 7:143. https://doi.org/10.1186/s40168-019-0757-8
Garcia MO, Templer PH, Sorensen PO, Sanders-DeMott R, Groffman PM, Bhatnagar JM (2020) Soil Microbes Trade-Off Biogeochemical Cycling for Stress Tolerance Traits in Response to Year-Round Climate Change. Front Microbiol 11:616. https://doi.org/10.3389/fmicb.2020.00616
Guo L, Zheng S, Cao C, Li C (2016) Tillage practices and straw-returning methods affect topsoil bacterial community and organic C under a rice-wheat cropping system in central China. Sci Rep 6:33155. https://doi.org/10.1038/srep33155
Hussain S, Khaliq A, AliHafiz B, Hussain A, Qadir T, Hussain S (2019) Temperature Extremes: Impact on Rice Growth and Development. In: Plant Abiotic Stress Tolerance, pp 153–171
Jansson JK, Hofmockel KS (2020) Soil microbiomes and climate change. Nat Rev Microbiol 18:35–46. https://doi.org/10.1038/s41579-019-0265-7
Jia X, Zhang N, Zhao Y, Wang L, Zhang C, Li X, Cao K, Gao Y (2020) A consecutive 4-year elevated air temperature shaped soil bacterial community structure and metabolic functional groups in the rhizosphere of black locust seedlings exposed to lead pollution. Sci Total Environ 732:139273. https://doi.org/10.1016/j.scitotenv.2020.139273
Jin T, Wang Y, Huang Y, Xu J, Zhang P, Wang N, Liu X, Chu H, Liu G, Jiang H et al (2017) Taxonomic structure and functional association of foxtail millet root microbiome. Gigascience 6. https://doi.org/10.1093/gigascience/gix089
Koyama A, Steinweg JM, Haddix ML, Dukes JS, Wallenstein MD (2018) Soil bacterial community responses to altered precipitation and temperature regimes in an old field grassland are mediated by plants. FEMS Microbiol Ecol 94:fix156. https://doi.org/10.1093/femsec/fix156
Kwak M-J, Kong HG, Choi K, Kwon S-K, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ et al (2018) Rhizoshere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36:1100-+. https://doi.org/10.1038/nbt.4232
Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J et al (2009) The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci U S A 106:15527–15533. https://doi.org/10.1073/pnas.0903507106
Legendre P (2014) Interpreting the replacement and richness difference components of beta diversity. Global Ecol Biogeogr 23:1324–1334. https://doi.org/10.1111/geb.12207
Liu Y, Li M, Zheng J, Li L, Zhang X, Zheng J, Pan G, Yu X, Wang J (2014) Short-term responses of microbial community and functioning to experimental CO2 enrichment and warming in a Chinese paddy field. Soil Biol Biochem 77:58–68. https://doi.org/10.1016/j.soilbio.2014.06.011
Liu Y, Zhang H, Xiong MH, Li F, Li LQ, Wang GL, Pan GX (2017) Abundance and composition response ofwheat field soil bacterial and fungal communities to elevated CO2 and increased air temperature. Biol Fertil Soils 53:3–8. https://doi.org/10.1007/s00374-016-1159-8
Mei R, Nobu MK, Liu WT (2020) Identifying anaerobic amino acids degraders through the comparison of short‐term and long‐term enrichments. Env Microbiol Rep 12:173–184. https://doi.org/10.1111/1758-2229.12821
Minemba D, Martin BC, Ryan MH, Veneklaas EJ, Gleeson DB (2020) Phosphate fertiliser alters carboxylates and bacterial communities in sweet potato (Ipomoea batatas(L.) Lam.) rhizosheaths. Plant Soil 454:245–260. https://doi.org/10.1007/s11104-020-04646-6
Mori H, Maruyama F, Kato H, Toyoda A, Dozono A, Ohtsubo Y, Nagata Y, Fujiyama A, Tsuda M, Kurokawa K (2014) Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res 21:217–227. https://doi.org/10.1093/dnares/dst052
Padhy SR, Bhattacharyya P, Nayak AK, Dash PK, Roy KS, Baig MJ, Mohapatra T (2020) Key Metabolic Pathways of Sulfur Metabolism and Bacterial Diversity under Elevated CO2 and Temperature in Lowland Rice: A Metagenomic Approach. Geomicrobiol J 37:13–21. https://doi.org/10.1080/01490451.2019.1657992
Philippot L, Spor A, Henault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron PA (2013) Loss in microbial diversity affects nitrogen cycling in soil. ISME J 7:1609–1619. https://doi.org/10.1038/ismej.2013.34
Pii Y, Borruso L, Brusetti L, Crecchio C, Cesco S, Mimmo T (2016) The interaction between iron nutrition, plant species and soil type shapes the rhizosphere microbiome. Plant Physiol Bioch 99:39–48. https://doi.org/10.1016/j.plaphy.2015.12.002
Raoufi RS, Soufizadeh S (2020) Simulation of the impacts of climate change on phenology, growth, and yield of various rice genotypes in humid sub-tropical environments using AquaCrop-Rice. Int J Biometeorol 64:1657–1673. https://doi.org/10.1007/s00484-020-01946-5
Reinhold-Hurek B, Bunger W, Burbano CS, Sabale M, Hurek T (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53:403–424. https://doi.org/10.1146/annurev-phyto-082712-102342
Ren G, Zhu C, Alam MS, Tokida T, Sakai H, Nakamura H, Usui Y, Zhu J, Hasegawa T, Jia Z (2015) Response of soil, leaf endosphere and phyllosphere bacterial communities to elevated CO2 and soil temperature in a rice paddy. Plant Soil 392:27–44. https://doi.org/10.1007/s11104-015-2503-8
Rinnan R, Michelsen A, Baath E, Jonasson S (2007) Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Global Change Biol 13:28–39. https://doi.org/10.1111/j.1365-2486.2006.01263.x
Sanguin H, Sarniguet A, Gazengel K, Moenne-Loccoz Y, Grundmann GL (2009) Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol 184:694–707. https://doi.org/10.1111/j.1469-8137.2009.03010.x
Santos-Medellin C, Edwards J, Liechty Z, Bao N, Sundaresan V (2017) Drought Stress Results in a Compartment-Specific Restructuring of the Rice Root-Associated Microbiomes. Mbio 8:e00764–00717. https://doi.org/10.1128/mBio.00764-17
Sasse J, Martinoia E, Northen T (2018) Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci 23:25–41. https://doi.org/10.1016/j.tplants.2017.09.003
Singh V, Sharma S, Shukla KP (2012) Harnessing Rhizospheric Bacterial Diversity of Important Medicinal Plants of Central India for Bioprospecting. In Vitro Cell Dev-An 48:79.
Stopnisek N, Shade A (2021) Persistent microbiome members in the common bean rhizosphere: an integrated analysis of space, time, and plant genotype. ISME J 15:2708–2722. https://doi.org/10.1038/s41396-021-00955-5
Szoboszlay M, Naether A, Mitterbauer E, Bender J, Weigel H-J, Tebbe CC (2017) Response of the rhizosphere prokaryotic community of barley (Hordeum vulgare L.) to elevated atmospheric CO2 concentration in open-top chambers. Microbiologyopen 6:e462. https://doi.org/10.1002/mbo3.462
Trivedi P, Anderson IC, Singh BK (2013) Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol 21:641–651. https://doi.org/10.1016/j.tim.2013.09.005
Ueda Y, Frindte K, Knief C, Ashrafuzzaman M, Frei M (2016) Effects of Elevated Tropospheric Ozone Concentration on the Bacterial Community in the Phyllosphere and Rhizoplane of Rice. Plos One 11:e0163178. https://doi.org/10.1371/journal.pone.0163178
Ullah A, Akbar A, Luo QQ, Khan AH, Manghwar H, Shaban M, Yang XY (2019) Microbiome Diversity in Cotton Rhizosphere Under Normal and Drought Conditions. Microb Ecol 77:429–439. https://doi.org/10.1007/s00248-018-1260-7
Wang XL, Wang MX, Xie XG, Guo SY, Zhou Y, Zhang XB, Yu N, Wang ET (2020) An amplification-selection model for quantified rhizosphere microbiota assembly. Sci Bull 65:983–986. https://doi.org/10.1016/j.scib.2020.03.005
Xiao D, Liu X, Yang R, Tan Y, Zhang W, He X, Xu Z, Wang K (2020) Nitrogen fertilizer and Amorpha fruticosa leguminous shrub diversely affect the diazotroph communities in an artificial forage grassland. Sci Total Environ 711:134967. https://doi.org/10.1016/j.scitotenv.2019.134967
Xiong L, Liu X, Vinci G, Spaccini R, Drososa M, Li L, Piccolo A, Pan G (2019) Molecular changes of soil organic matter induced by root exudates in a rice paddy under CO2 enrichment and warming of canopy air. Soil Biol Biochem 137:107544. https://doi.org/10.1016/j.soilbio.2019.107544
Xu J, Zhang Y, Zhang P, Trivedi P, Riera N, Wang Y, Liu X, Fan G, Tang J, Coletta-Filho HD et al (2018) The structure and function of the global citrus rhizosphere microbiome. Nat Commun 9:4894. https://doi.org/10.1038/s41467-018-07343-2
Yan Y, Kuramae EE, de Hollander M, Klinkhamer PGL, van Veen JA (2017) Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J 11:56–66. https://doi.org/10.1038/ismej.2016.108
Yao MJ, Rui JP, Niu HS, Heděnec P, Li JB, He ZL, Wang JM, Cao WD, Li XZ (2017) The differentiation of soil bacterial communities along a precipitation and temperature gradient in the eastern Inner Mongolia steppe. Catena 152:47–56. https://doi.org/10.1016/j.catena.2017.01.007
Yin C, Vargas JMC, Schlatter DC, Hagerty CH, Hulbert SH, Paulitz TC (2021) Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome 9. https://doi.org/10.1186/s40168-020-00997-5
Yu X, Liu X, Liu X (2019) Response of rhizosphere bacterial community of Taxus chinensis var. mairei to temperature changes. Plos One 14:e0226500. https://doi.org/10.1371/journal.pone.0226500
Zeng J, Shen JP, Wang JT, Hu HW, Zhang CJ, Bai R, Zhang LM, He JZ (2018) Impacts of Projected Climate Warming and Wetting on Soil Microbial Communities in Alpine Grassland Ecosystems of the Tibetan Plateau. Microb Ecol 75:1009–1023. https://doi.org/10.1007/s00248-017-1098-4
Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi Sj, Cho H, Karaoz U, Loque D, Bowen BP et al (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480. https://doi.org/10.1038/s41564-018-0129-3
Zhang J, Liu Y-X, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X et al (2019a) NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37:676-+. https://doi.org/10.1038/s41587-019-0104-4
Zhang Y, Dong SK, Gao QZ, Liu SL, Zhou HK, Ganjurjav H, Wang XX (2016) Climate change and human activities altered the diversity and composition of soil microbial community in alpine grasslands of the Qinghai-Tibetan Plateau. Sci Total Environ 562:353–363. https://doi.org/10.1016/j.scitotenv.2016.03.221
Zhang Y, Gao X, Shen Z, Zhu C, Jiao Z, Li R, Shen Q (2019b) Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant Soil 439:553–567. https://doi.org/10.1007/s11104-019-04055-4
Zhang Y, Jiang W, Li Q, Xu W, Wang J, Hu J, Zhang Z (2021) Soil nutrient levels determine the variation of bacterial communities in the rhizosphere of rice under different conditions of climate and genotype. Appl Soil Ecol 167:104025. https://doi.org/10.1016/j.apsoil.2021.104025
Zhou J, Jiang X, Wei D, Zhao B, Ma M, Chen S, Cao F, Shen D, Guan D, Li J (2017) Consistent effects of nitrogen fertilization on soil bacterial communities in black soils for two crop seasons in China. Sci Rep 7:3267. https://doi.org/10.1038/s41598-017-03539-6
Zhu C, Ling N, Li L, Liu X, Dippold MA, Zhang X, Guo S, Kuzyakov Y, Shen Q (2020) Compositional variations of active autotrophic bacteria in paddy soils with elevated CO2 and temperature. Soil Ecol Lett 2:295–307. https://doi.org/10.1007/s42832-020-0044-4
Funding
This work was supported by the National Key Research and Development Plan of China (2016YFD0300502), the National Natural Science Foundation of China (41977053) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that there is no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
All authors agree to publish the paper.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Zhang, Y., Xu, W. et al. Possible effects of temperature on bacterial communities in the rhizosphere of rice under different climatic regions. Arch Microbiol 204, 212 (2022). https://doi.org/10.1007/s00203-022-02812-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02812-1