Abstract
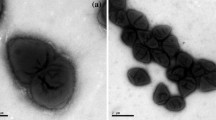
A Gram-stain-negative, pink-pigmented, aerobic, non-motile and rod-shaped bacterium, designated as strain H15T, was isolated from Acmaea sp., collected from Weihai, Shandong Province, China. The novel isolate was able to grow at 4–37 °C (optimum 33 °C), pH 5.5–9.0 (optimum 7.0) and with 0.0–7.0% NaCl (optimum 4%, w/v). Phylogenetic analysis based on 16S rRNA gene sequence comparisons revealed that the strain belonged to the family Rhodobacteraceae and was associated to the type strain of Pseudaestuariivita atlantica (96.7%). Genome analysis showed that the genome size was 3,893,398 bp and the DNA G + C content obtained from the draft genome sequence was 56.7%. The secondary metabolites predicated that the strain H15T contained one cluster of lasso peptide, one cluster of bacteriocin, two clusters of terpene production, two clusters of homoserine lactone and one cluster of beta lactone. The average amino acid identity, average nucleotide identity and digital DNA–DNA hybridization values between genome sequences of strain H15T and all the related strains compared were lower than 63.1, 72.0 and 19.7%, respectively. Based on the analysis of chemical components, the predominant cellular fatty acids were summed featured 8 (C18:1ω7c/ω6c, 46.1%), C20:1 ω7c (17.1%), the major polar lipids contained phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine and an unidentified lipid and the predominant menaquinone was Q10. Therefore, the combined chemotaxonomic, phenotypic and phylogenetic data indicated that the strain was considered to represent a novel species of the genus Pseudaestuariivita and the name Pseudaestuariivita rosea sp. nov. was proposed for strain H15T (MCCC 1K04420T = KCTC 82505T).

Similar content being viewed by others
Abbreviations
- AAI:
-
Average amino acid identity
- ANI:
-
Average nucleotide identity
- cAAI:
-
Core-gene average amino acid identity
- COG:
-
Cluster of orthologous group
- dDDH:
-
Digital DNA–DNA hybridization;
- GGDC:
-
Genome-to-genome distance calculator
- HPLC:
-
High performance liquid chromatography
- KCTC:
-
Korean collection for type cultures
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LPS:
-
Lipopolysaccharide
- MA:
-
Marine agar 2216
- MB:
-
Marine broth 2216
- MCCC:
-
Marine culture collection of China
- MEGA:
-
Molecular evolutionary genetics analysis
- NCBI:
-
National Center for Biotechnology Information
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- POCP:
-
Percent of conserved proteins
- TLC:
-
Two-dimensional thin-layer chromatogram
- VFDB:
-
Virulence factors of pathogenic bacteria
References
Aziz RK, Bartels D, Best AA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Blin K, Shaw S, Steinke K et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/nar/gkz310
Bowman J (2000) Description of Cellulophaga algicola sp. Nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb nov. Int J Syst Evol Microbiol 50(5):1861–1868. https://doi.org/10.1099/00207713-50-5-1861
Chen L, Yang J, Yu J et al (2005) VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. https://doi.org/10.1093/nar/gki008
Chun J, Oren A, Ventosa A et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Consortium TU (2018) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. https://doi.org/10.1093/nar/gky1049
Felsenstein J (1981) Evolutionary trees from DNA sequences: a Maximum Likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Hiraishi A, Ueda Y, Ishihara J et al (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469. https://doi.org/10.2323/jgam.42.457
Holden-Dye K, Crouch LI, Jones MR (2008) Structure, function and interactions of the PufX protein. Biochim Biophys Acta 1777:613–630. https://doi.org/10.1016/j.bbabio.2008.04.015
Kanehisa M, Sato Y, Kawashima M et al (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. https://doi.org/10.1093/nar/gkv1070
Kim M, Oh HS, Park SC et al (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. https://doi.org/10.1099/ijs.0.059774-0
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Konstantinidis KT, Tiedje JM (2005) Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. https://doi.org/10.1128/jb.187.18.6258-6264.2005
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Li GZ, Lai QL, Du YP et al (2015) Aestuariivita atlantica sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol 65:3281–3285. https://doi.org/10.1099/ijsem.0.000406
Meier-Kolthoff GM (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff AAF, Klenk HP et al (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) Fatty acid, polar lipid and wall amino acid composition of Gardnerella vaginalis. Arch Microbiol 138:68–71. https://doi.org/10.1007/BF00425410
Park S, Won SM, Kim H et al (2014) Aestuariivita boseongensis gen. nov., sp. nov., isolated from a tidal flat sediment. Int J Syst Evol Microbiol 64:2969–2974. https://doi.org/10.1099/ijs.0.062406-0
Rodriguez-R L, Konstantinidis K (2014) Bypassing cultivation to identify bacterial species: culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe. https://doi.org/10.1128/microbe.9.111.1
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sawant SS, Salunke BK, Kim BS (2015) A rapid, sensitive, simple plate assay for detection of microbial alginate lyase activity. Enzyme Microb Technol 77:8–13. https://doi.org/10.1016/j.enzmictec.2015.05.003
Senghor B, Bassene H, Khelaifia S et al (2019) Oceanobacillus timonensis sp. nov. and Oceanobacillus senegalensis sp. nov., two new moderately halophilic, Gram-stain positive bacteria isolated from stools sample of healthy young Senegalese. Antonie Van Leeuwenhoek 112:785–796. https://doi.org/10.1007/s10482-018-01212-0
Smibert RM, Krieg NR (1994) Phenotypic characterization: methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Tindall BJ, Sikorski J, Smibert R et al (2007) Phenotypic charaterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM et al (eds) Methods for general and molecular microbiology, pp 330–393. https://doi.org/10.1128/9781555817497.ch15
Tippmann HF (2004) Analysis for free: comparing programs for sequence analysis. Brief Bioinform 5(1):82–87. https://doi.org/10.1093/bib/5.1.82
Wirth JS, Whitman WB (2018) Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int J Syst Evol Microbiol 68:2393–2411. https://doi.org/10.1099/ijsem.0.002833
Yang SJ, Cho JC (2008) Gaetbulibacter marinus sp nov., isolated from coastal seawater, and emended description of the genus Gaetbulibacter. Int J Syst Evol Microbiol 58:315–318. https://doi.org/10.1099/ijs.0.65382-0
Yin R, Yi YJ, Chen Z et al (2021) Flavihalobacter algicola gen. nov. sp. nov., a member of the family Flavobacteriaceae with alginate-degradation activity, isolated from marine alga Saccharina japonica. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.004701
Yoon SH, Ha SM, Kwon SJ et al (2017a) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Yoon SH, Ha SM, Lim J et al (2017b) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Acknowledgements
We thank Prof. Aharon Oren for his valuable help with naming the species.
Funding
This work was supported by the National Natural Science Foundation of China (31700116), the Natural Science Foundation of Shandong Province (ZR2017MC019), the China Postdoctoral Science Foundation (2017M62218), and the Key Science and Technology Program of Weihai (1070413421511).
Author information
Authors and Affiliations
Contributions
YJY contributed to the sample collection and biochemical characterization. STY analysed the data and drafted the manuscript. ZC contributed to the construction of phylogenetic tree. MJZ contributed to revision of this article. SKG contributed to genome submission and analyses. YXZ designed the experiments and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yun, ST., Chen, Z., Yi, YJ. et al. Pseudaestuariivita rosea sp. nov., isolated from Acmaea sp., a marine mollusk. Arch Microbiol 204, 94 (2022). https://doi.org/10.1007/s00203-021-02690-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-021-02690-z